Abstract
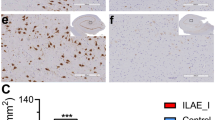
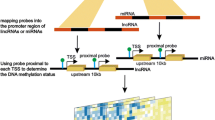
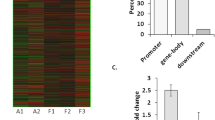
Synaptic protein shanks (SH3 and multiple ankyrin repeat domains protein, Shank) have emerged as an important mediator of synaptic remodeling. Synaptic remodeling is a common pathogenic process in various neurological disorders including epilepsy. However, the expression and function of shanks gene in epileptogenesis has not been investigated to date. Herein, we investigated the expression of shanks (shank1/2/3) mRNA expression in both epileptic rats and epilepsy patients. Furthermore, methyl target sequencing was applied to explore the relationship between shank mRNA expression and DNA methylation in both rats and human patients. In general rat model, shank1/2/3 mRNA was downregulated at acute stage, upregulated at latent stage, and returned to the basal level at chronic stage. Ten CpG sites of shank1 was found differentially methylated, out of which 6 were hypermethylated. Seventeen CpG sites of shank3 were differentially methylated, out of which 8 were hypermethylated. In human epilepsy patients, decreased shank2 mRNA was detected from the brain tissue, with DNA hypermethylation dominant from both brain (18 out of 30) and blood tissue (58 out of 80), indicating the regulation role of DNA methylation on shank2 expression. In conclusion, our finding suggests the participation of the shanks gene in the pathophysiology of seizure, out of which 2 shank3 CpG sites (Chr7: 130473419, and Chr7: 130473405) may play an important role in shank3 expression at both the acute and latent stages in the SE rat model.






Similar content being viewed by others
References
Keller R, Basta R, Salerno L, Elia M (2017) Autism, epilepsy, and synaptopathies: a not rare association. Neurol Sci 38:1353–1361
Krishnan V, Stoppel DC, Nong Y, Johnson MA, Nadler MJS, Ozkaynak E, Teng BL, Nagakura I et al (2017) Autism gene Ube3a and seizures impair sociability by repressing VTA Cbln1. Nature 543:507–512
Abeliovich A, Gitler AD (2016) Defects in trafficking bridge Parkinson’s disease pathology and genetics. Nature 539:207–216
Lourenco MV, Frozza RL, de Freitas GB, Zhang H, Kincheski GC, Ribeiro FC, Gonçalves RA, Clarke JR et al (2019) Exercise-linked FNDC5/irisin rescues synaptic plasticity and memory defects in Alzheimer’s models. Nat Med 25:165–175
Karayannis T, Au E, Patel JC, Kruglikov I, Markx S, Delorme R, Héron D, Salomon D et al (2014) Cntnap4 differentially contributes to GABAergic and dopaminergic synaptic transmission. Nature 511:236–240
Soltesz I, Alger BE, Kano M, Lee SH, Lovinger DM, Ohno-Shosaku T, Watanabe M (2015) Weeding out bad waves: towards selective cannabinoid circuit control in epilepsy. Nat Rev Neurosci 16:264–277
Khlebodarova TM, Kogai VV, Trifonova EA, Likhoshvai VA (2018) Dynamic landscape of the local translation at activated synapses. Mol Psychiatry 23:107–114
Shin SM, Zhang N, Hansen J, Gerges NZ, Pak DTS, Sheng M, Lee SH (2012) GKAP orchestrates activity-dependent postsynaptic protein remodeling and homeostatic scaling. Nat Neurosci 15:1655–1666
Yuan T, Mameli M, O'Connor EC, Dey PN, Verpelli C, Sala C, Perez-Otano I, Lüscher C et al (2013) Expression of cocaine-evoked synaptic plasticity by GluN3A-containing NMDA receptors. Neuron 80:1025–1038
Monteiro P, Feng G (2017) SHANK proteins: roles at the synapse and in autism spectrum disorder. Nat Rev Neurosci 18:147–157
Berg EL, Copping NA, Rivera JK, Pride MC, Careaga M, Bauman MD, Berman RF, Lein PJ et al (2018) Developmental social communication deficits in the Shank3 rat model of Phelan-McDermid syndrome and autism spectrum disorder. Autism Res 11:587–601
de Sena Cortabitarte A, Degenhardt F, Strohmaier J, Lang M, Weiss B, Roeth R, Giegling I, Heilmann-Heimbach S et al (2017) Investigation of SHANK3 in schizophrenia. Am J Med Genet Part B Neuropsychiatr Genet 174:390–398
Machnes ZM, Huang TCT, Chang PKY, Gill R, Reist N, Dezsi G, Ozturk E, Charron F et al (2013) DNA methylation mediates persistent epileptiform activity in vitro and in vivo. PLoS One 8:e76299
Kobow K, Jeske I, Hildebrandt M, Hauke J, Hahnen E, Buslei R, Buchfelder M, Weigel D et al (2009) Increased reelin promoter methylation is associated with granule cell dispersion in human temporal lobe epilepsy. J Neuropathol Exp Neurol 68:356–364
Kobow K, Blümcke I (2011) The methylation hypothesis: do epigenetic chromatin modifications play a role in epileptogenesis? Epilepsia 52(Suppl 4):15–19
Campbell RR, Wood MA (2019) How the epigenome integrates information and reshapes the synapse. Nat Rev Neurosci 20:133–147
Beri S, Tonna N, Menozzi G, Bonaglia MC, Sala C, Giorda R (2007) DNA methylation regulates tissue-specific expression of Shank3. J Neurochem 101:1380–1391
Luo ZH, Walid A A, Xie Y, Long H, Xiao W, Xu L, Fu Y, Feng L et al (2019) Construction and analysis of a dysregulated lncRNA-associated ceRNA network in a rat model of temporal lobe epilepsy. Seizure 69:105–114
Fu Y, Yang MS, Jiang J, Ganesh T, Joe E, Dingledine R (2015) EP2 receptor signaling regulates microglia death. Mol Pharmacol 88:161–170
Xiao W, Cao Y, Long H, Luo Z, Li S, Deng N, Wang J, Lu X et al (2018) Genome-wide DNA methylation patterns analysis of noncoding RNAs in temporal lobe epilepsy patients. Mol Neurobiol 55:793–803
Long H-Y, Feng L, Kang J, Luo ZH, Xiao WB, Long LL, Yan XX, Zhou L et al (2017) Blood DNA methylation pattern is altered in mesial temporal lobe epilepsy. Sci Rep 7:43810
Xiao W, Wu Y, Wang J, Luo Z, Long L, Deng N, Ning S, Zeng Y et al (2019) Network and pathway-based analysis of single-nucleotide polymorphism of miRNA in temporal lobe epilepsy. Mol Neurobiol 56:7022–7031
Uchino S, Wada H, Honda S, Nakamura Y, Ondo Y, Uchiyama T, Tsutsumi M, Suzuki E et al (2006) Direct interaction of post-synaptic density-95/Dlg/ZO-1 domain-containing synaptic molecule Shank3 with GluR1 alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor. J Neurochem 97:1203–1214
Marwick K, Skehel P, Hardingham G, Wyllie D (2015) Effect of a GRIN2A de novo mutation associated with epilepsy and intellectual disability on NMDA receptor currents and Mg(2+) block in cultured primary cortical neurons. Lancet Lond Engl 385(Suppl 1):S65
Li D, Yuan H, Ortiz-Gonzalez XR, Marsh ED, Tian L, McCormick EM, Kosobucki GJ, Chen W et al (2016) GRIN2D recurrent de novo dominant mutation causes a severe epileptic encephalopathy treatable with NMDA receptor channel blockers. Am J Hum Genet 99:802–816
Rothan HA, Amini E, Faraj FL, Golpich M, Teoh TC, Gholami K, Yusof R (2017) NMDA receptor antagonism with novel indolyl, 2-(1,1-dimethyl-1,3-dihydro-benzo[e]indol-2-ylidene)-malonaldehyde, reduces seizures duration in a rat model of epilepsy. Sci Rep 7:45540
Chang P, Augustin K, Boddum K, Williams S, Sun M, Terschak JA, Hardege JD, Chen PE et al (2016) Seizure control by decanoic acid through direct AMPA receptor inhibition. Brain J Neurol 139:431–443
Yelshanskaya MV, Singh AK, Sampson JM, Narangoda C, Kurnikova M, Sobolevsky AI (2016) Structural bases of noncompetitive inhibition of AMPA-subtype Ionotropic glutamate receptors by antiepileptic drugs. Neuron 91:1305–1315
Peça J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z et al (2011) Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature 472:437–442
Mei Y, Monteiro P, Zhou Y, Kim JA, Gao X, Fu Z, Feng G (2016) Adult restoration of Shank3 expression rescues selective autistic-like phenotypes. Nature 530:481–484
Zhou Y, Kaiser T, Monteiro P, Zhang X, van der Goes MS, Wang D, Barak B, Zeng M et al (2016) Mice with Shank3 mutations associated with ASD and schizophrenia display both shared and distinct defects. Neuron 89:147–162
Funding
This work was supported through funding from the National Natural Science Foundation of China (grant numbers 81671299 to B.X., and 81701182 to Y.F.) and the Natural Science Foundation of Hunan Province (grant number: 2018JJ2648) to Z.L.
Author information
Authors and Affiliations
Contributions
Du Liu and Zhaohui Luo conducted the majority of the experiments. Yujiao Fu analyzed and interpreted the data and drafted the manuscript. Jialing Guo and Wei Xiao prepared the epilepsy animal experiment. Du Liu and Zhaohui Luo prepared the CpG detective experiments. Hongyu Long and Wenbiao Xiao analyzed and interpreted the data. Li Feng prepared for language editing and revising. Bo Xiao and Zhaohui Luo contributed to the study design. Bo Xiao provided financial support. All authors consent to the publication of the manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethics Approval
All procedures were followed in accordance with the ethical standards of the responsible committee on animal and human experimentation (the Ethics Review Committee of Xiangya Hospital, numbers 201603296 and 201603297) and with the Helsinki Declaration of 1964 and later versions.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Fu, Y., Liu, D., Guo, J. et al. Dynamic Change of Shanks Gene mRNA Expression and DNA Methylation in Epileptic Rat Model and Human Patients. Mol Neurobiol 57, 3712–3726 (2020). https://doi.org/10.1007/s12035-020-01968-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-020-01968-5