Abstract
For interpreting the pressure induced shifts of resonance lines of folded as well as unfolded proteins the availability of data from well-defined model systems is indispensable. Here, we report the pressure dependence of 1H and 15N chemical shifts of the side chain atoms in the protected tetrapeptides Ac-Gly-Gly-Xxx-Ala-NH2 (Xxx is one of the 20 canonical amino acids) measured at 800 MHz proton frequency. As observed earlier for other nuclei the chemical shifts of the side chain nuclei have a nonlinear dependence on pressure in the range from 0.1 to 200 MPa. The pressure response is described by a second degree polynomial with the pressure coefficients B1 and B2 that are dependent on the atom type and type of amino acid studied. A number of resonances could be assigned stereospecifically including the 1H and 15N resonances of the guanidine group of arginine. In addition, stereoselectively isotope labeled SAIL amino acids were used to support the stereochemical assignments. The random-coil pressure coefficients are also dependent on the neighbor in the sequence as an analysis of the data shows. For Hα and HN correction factors for different amino acids were derived. In addition, a simple correction of compression effects in thermodynamic analysis of structural transitions in proteins was derived on the basis of random-coil pressure coefficients.
Similar content being viewed by others
Introduction
The study of the pressure response of polypeptides and proteins by high pressure NMR spectroscopy can be used for characterization of the free energy landscape of proteins (for reviews see e.g. Kitahara et al. 2013; Akasaka and Matsuki 2015). The pressure response allows the detection of rare “excited” conformational states of proteins that are important for folding and function (see e.g. Kalbitzer et al. 2009). Excited states also provide the basis for the development of a new type of allosteric inhibitors of proteins involved in signal transduction, called intrinsic allosteric inhibitors (Rosnizeck et al. 2010, 2012; Kalbitzer et al. 2013a; Kalbitzer and Spoerner b).
For the detection of rare states of proteins, mainly pressure dependent changes of chemical shifts are evaluated that are in many cases non-linear and can often be fitted with an appropriate thermodynamical model. These conformational contributions have to be separated from other chemical shift contributions as they are observed even in non-folded model compounds as a consequence of compression and rearrangement of the water shell. A non-linear pressure response of chemical shifts can even be determined experimentally in the peptide bond model N-methyl acetamide and is supported by quantum chemical calculations (Frach et al. 2016). However, random-coil peptides are better suited models for protein work. Arnold et al. (2002) reported the first data set for the main chain and side chain protons of the tetrapeptide Gly-Gly-Xxx-Ala in the pressure range up to 200 MPa. The pressure dependence of chemical shifts δ can sufficiently well be described by a second order polynomial with the chemical shift at pressure P0 (0.1 MPa) and the first and second order pressure coefficients B1 and B2. A data set recorded at 800 MHz was also published for all backbone atoms and the carbon resonances of the side chains of the protected tetrapeptide Ac-Gly-Gly-Xxx-Ala-NH2 (Koehler et al. 2012; Beck Erlach et al. 2016, 2017).
The quality of data by Arnold et al. (2002) measured at 600 MHz was not sufficient to determine the second order coefficient for the side chain protons. In this paper, we will present such data recorded at the N- and C-terminally protected tetrapeptide together with the pressure response of nitrogen side chain atoms not yet reported. With these data a complete data set for all nuclei of the model tetrapeptide Ac-Gly-Gly-Xxx-Ala-NH2 will be available for the scientific community.
Materials and methods
Synthesis of tetrapeptides
Uniformly 13C and 15N enriched and Fmoc (N-(9-Fluorenylmethoxycarbonyl)) protected amino acids required for the synthesis were purchased from Sigma Aldrich (St. Louis, MO, USA). The isotope enrichment is larger than 98%. All other chemicals were purchased from Merck (Darmstadt, Germany).
The synthesis of the tetrapeptide Ac-Gly-Gly-Xxx-Ala-NH2, where Xxx stands for one of the 20 canonical amino acids, was already described in detail earlier (Koehler et al. 2012; Beck Erlach et al. 2016). Only the amino acid Xxx at position 3 in the tetrapeptide Ac-Gly-Gly-Xxx-Ala-NH2 was uniformly 13C and 15N enriched. The purity of the tetrapeptides was confirmed by ESI–MS (Bruker, Billerica, MA, USA) and RP-HPLC (Waters, Milford, MA, USA).
Selectively enriched SAIL amino acids
Stereoselectively 2H, 15N and 13C labeled amino acids were obtained from SAIL Technologies Inc. (Tokyo, Japan). For more details, see Kainosho et al. (2006), Kainosho and Güntert (2009).
Sample preparation
Tetrapeptide samples were prepared by dissolving 2.5 µmol of the peptide in 500 µL of a buffer containing 20 mM perdeuterated Tris-d11 (Tris(hydroxymethyl-d3)amino-d2-methane) and 0.5 mM DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid) with a ratio H2O:D2O of 90:10. Thus a final peptide concentration of 5 mM was obtained. The pH value was adjusted to 6.7 using a Hamilton Spintrode attached to a Beckman Coulter pH meter. Histidine was also measured at pH 4.0 and pH 8.5. The pH values have not been corrected for the deuterium isotope effect.
For the stereospecific assignment of amino acids, unlabeled amino acids or (stereo)-selectively 2H, 13C enriched SAIL amino acids were dissolved in 20 mM Tris-d11, 0.5 mM DSS and 10% D2O, pH 6.7, to obtain a final amino acid concentration of 4 to 10 mM.
NMR spectroscopy
Most of the experiments were performed on an 800 MHz Bruker Avance spectrometer (Bruker, Billerica, MA, USA) with a room temperature probe head (QXI). The experiments were performed at 283 K, with a temperature calibration carried out after each sample change by measuring the difference of the proton resonance of the hydroxyl and the methyl group in 100% methanol as described by Raiford et al. (1979).
1H-NMR spectra were directly referenced to the methyl resonances of internal DSS, 15N signals were indirectly referenced to DSS using a Ξ-value of 0.101329118 (15N/1H) (Wishart et al. 1995b). Atom labels were named according to IUPAC recommendations (Markley et al. 1998).
1H and 15N chemical shifts were obtained from highly resolved 1D proton and 2D [1H, 15N]-HSQC spectra with a typical digital resolution of the time domain data of 0.04 Hz (1H) and 0.32 Hz (15N). A Lorentzian-to-Gaussian transformation was applied to the FID to obtain signals as narrow as possible.
Data acquisition and processing was performed with Bruker TopSpin 3.2 PL6. For peak picking the software AUREMOL (Gronwald and Kalbitzer, 2004) was used. Data evaluation and fitting was done with the software package R (R Core Team 2019).
High pressure system
The high pressure system, especially the autoclave holding the ceramic cell was described in detail by Koehler et al. (2012). Pressure was applied to the NMR sample via pressurized fluids (methylcyclohexane or water) contained in high pressure lines. For generating the pressure a manually operated piston compressor and an air-to-liquid pressure intensifier (Barocycler®, HUB440, Pressure BioSciences Inc., South Easton, MA, USA), which is controlled by the spectrometer, were used. The ceramic cell was purchased from Daedalus Innovations LLC (Aston, PA, USA) with a maximum pressure limit of 250 MPa. For safety reasons pressure was only applied up to 200 MPa. The autoclave holding the ceramic cell is similar to the original autoclave (Peterson and Wand 2005) provided by Daedalus Innovations LLC but has an integrated safety valve, similar to the security valve described by Beck Erlach et al. (2010).
Data evaluation
For all 20 model peptides a pressure series from 1 to 200 MPa was performed. The obtained pressure dependent chemical shifts δ were fitted as a function of pressure P with a second degree polynomial
with P0 the atmospheric pressure of 0.1 MPa and δ0 the chemical shift at pressure P0. B1 and B2 are the first and second order pressure coefficients. For random coil peptides, a second degree polynomial as defined by Eq. 1 is sufficient in the pressure range studied here (Arnold et al. 2002; Koehler et al. 2012; Beck Erlach et al. 2016, 2017). The description of the pressure response of small molecules such as the GTP analog GTPγS sometimes requires a third degree polynomial (Spoerner et al. 2017). When the ratio of B2/B1 is negative, an extremum with slope zero would be reached at a pressure Pex with
An extremum has not observed for our model peptides in the range up to 200 MP. With the mean value of B2/B1 for Hβ (Table 2) of − 1.4 GPa−1 the maximum value would be expected at 375 MPa, far outside the pressure range studied. The dependence of the chemical shifts δ on pressure leads to an expression that is dependent on tanh (ΔG/2RT) (Beck Erlach et al. 2014). However, in a two state-model an extremum at high pressures is not expected from a thermodynamic description when ΔG is only linearly dependent on pressure P. This is the case when ΔG does not contain a second degree term, since he compressibility difference Δβ′ = -\(\frac{\partial {\Delta V}_{ij}^{0}}{\partial P}=\) 0 as most authors assume in their data evaluation. The tanh function can be described by a second order differential equation typical for physical processes that show a saturation like behavior (Kepner 2010). In contrast to our second degree polynomial, it shows an asymptotic behavior at high pressures that we call saturation-like in the following. In fact, the Taylor-series of tanh itself has a second order term of zero, therefore the second degree polynomial that is traditionally used for a fit of the data is not suitable for the description of two-state equilibrium with ΔG only linearily dependent on P. However, when Δβ′ is not zero, again a maximum is obtained and a second degree term is required for a proper description of the data.
Sequence dependent corrections for the pressure coefficients
Analogously to the method proposed by Schwarzinger et al. (2001) for the correction of the random-coil chemical shifts of atom a in amino acid x in position i by amino acid y in position i + j correction factors \({C}_{\mathrm{1,2}}^{-1,a}\)(y), \({C}_{\mathrm{1,2}}^{+1,a}\)(y), and \({C}_{\mathrm{1,2}}^{+2,a}\)(y) for the pressure coefficients B1 and B2 were calculated. They can be obtained from the pressure response of Gly1, Gly2, and Ala4 by subtracting the B1 and B2 values obtained for Ac-GGGA-NH2 from those obtained for Ac-GGyA-NH2. The correction factors \({C}_{\mathrm{1,2}}^{-1,a}\)(y) for the atom a (HN or Hα) of amino acid x in position i by amino acid y in position i-1 are given by
with \({B}_{\mathrm{1,2}}^{a}\) the pressure coefficients of atom a in Ala4. Analogously, the correction factors \({C}_{\mathrm{1,2}}^{+1,a}\)(y), and \({C}_{\mathrm{1,2}}^{+2,a}\)(y) are obtained from Gly2 and Gly1, respectively. The sequence corrected B1 and B2 values \({B}_{\mathrm{1,2}}^{a,corr}\) for atom a in amino acid x in the sequence -uxyz- are than given as
Results and discussion
Assignment of resonance lines
By applying pressure to the tetrapeptides and fitting the resulting pressure dependence of the chemical shift to Eq. 1, we obtained a complete dataset of 1H and 15N random coil chemical shift values for side chains of the amino acid 3 in the model peptides Ac-Gly-Gly-Xxx-Ala-NH2. The assignments of most 1H-resonances could be done on the basis of the already published proton assignments from Bundi and Wüthrich (1979) of Gly-Gly-Xxx-Ala and an analysis of the multiplet patterns. When necessary classical two-dimensional COSY, TOCSY and NOESY spectra were recorded. The 15N resonances could be assigned by HSQC-spectra from the adjacent protons already assigned.
Stereospecific assignments
Geminal proton resonances in stereo selectively labeled SAIL amino acids
At 800 MHz proton resonance frequency most of the geminal proton in the tetrapeptides are resolved and can be observed separately. The typical geminal coupling constant in sp3 bonds is -12 Hz, corresponding to 0.015 ppm at 800 MHz. For most methylene protons the peak separation is significantly larger. They can be observed separately but still strong coupling effects are visible (see below). This is also true for the isolated amino acids (Table 1). Since stereo selectively deuterated SAIL amino acids (Kainosho et al. 2006; Kainosho and Güntert 2009) are available, at least in the amino acids the geminal proton resonances can unequivocally be assigned. When comparing the spectra of isotope labeled with the corresponding unlabeled amino acids, a difficulty is the isotope shift that occurs in the SAIL amino acids since these are not only stereoselectively deuterated but also 15N and 13C enriched. In general, these isotope shifts are smaller than the separation of the corresponding proton resonances. The isotope labelling induces an upfield shift of the order of 0.014 to 0.043 ppm (Table 1). Since deuteration is not 100% complete because of the limited purity of the starting materials, for many geminal protons a weak signal of the unlabeled group can be detected (values in brackets in Table 1). But even here an upfield isotope shift is observed caused by the other nuclei (especially also the 13C-nuclei of the methylene groups). When the resonances of geminal protons are separated, the Hβ2 resonances are always shifted upfield relative to the Hβ3 resonances. The only exception is cysteine. These relative shifts also apply for the methylene resonances in γ-position of Ile and Met. However, for the Hγ2 and Hγ3 resonances of Lys and the Hδ2 and Hδ3 resonances of Pro the order is interchanged. These assignments give also a hint to the stereospecific assignments in the protected tetrapeptides (Table 2) but cannot prove their stereospecific assignment definitively.
Stereochemical assignment of Hβ-proton resonances in Ac-GGXA-NH2
Also in the protected tetrapeptides, a large number of geminal proton resonances are non-equivalent at 800 MHz proton resonance frequency and can be observed separately (Table 2). However, in literature most of them were not assigned stereospecifically, since the high internal mobility in random coil peptides does complicate the use of simple NMR methods developed for folded proteins that are based on the assumption of a single conformer in solution. Here, only a combination of 2D-NOESY-spectroscopy with extended molecular dynamic runs allows a safe assignment, as previously reported by Harsch et al. (2013) for GGNA-NH2 and GGQA-NH2. Although our tetrapeptides are additionally acetylated at N-terminus, these assignments can safely be transferred to our peptides since the N-terminal protection has only minor effects on the chemical shifts of the β-methylene groups (GGNA-NH2: 2.84 ppm, 2.75 ppm; Ac-GGNA-NH2: 2.84 ppm, 2.75 ppm; GGQA-NH2: 2.12 ppm, 1.99 ppm; Ac-GGQA-NH2; 2.13 ppm, 1.99 ppm). However, the SAIL data do not further corroborate these assignments. The Hβ2 resonances of glutamine are not separated in the free amino acid glutamine. The Hβ2 resonance in Ac-GGNA-NH2 has the same chemical shift as in the isolated amino acid (2.84 ppm) but instead of being upfield shifted the Hβ3 resonance is strongly downfield shifted by 0.19 ppm (Tables 1 and 2), indicating that other factors influencing the chemical shifts play a role here. A comparison of Hβ chemical shifts in the tetrapeptide and the SAIL amino acids allows a tentative stereochemical assignment of the resonances in Asp, Glu, His, Leu, Met, Phe, Pro, and Tyr. However, one has to be careful when using this assignment method since the peak separation is not very large: In Asn, one would have ended up with the wrong decision by just using the order of the chemical shifts (see above) in the free amino acid.
Methyl groups of Val and Leu
The stereochemical assignments of the methyl carbon resonances of Leu and Val in our tetrapeptides by selective deuteration were reported earlier by Beck Erlach et al. (2017). The assignment of corresponding proton resonances can be made directly from these data (see Table 3).
Amide and amino groups of Asn, Gln, and Arg
The stereochemical assignments of the side chain and C-terminal amide groups of Asn and Gln were earlier reported for the C-terminal protected tetrapeptides GGNA-NH2 and GGQA-NH2 by Harsch et al. (2013). As already stated above, our tetrapeptides are additionally acetylated at N-terminus. These assignments can safely transferred since the N-terminal protection has only minor effects on the chemical shifts. For the amide groups of GGNA-NH2 and Ac-GGNA-NH2 the shifts are (7.65, 6.96) ppm and (7.69, 6.99) ppm, respectively. The corresponding values of GGQA-NH2 and Ac-GGQA-NH2 are (7.59, 6.90) ppm and (7.64, 6.94) ppm, respectively. In fact, a general analysis of the BMRB data base shows that also in folded proteins the downfield shifted resonance lines can be assigned to Hδ21 and Hε21, with a separation of the chemical shifts of the two amide resonance lines s ≥ 0.40 ppm for asparagine and ≥ 0.42 ppm for glutamine, at a confidence level > 95% (Harsch et al. 2017). In the past, the proton and nitrogen resonances of the guanidino group of Arg in the tetrapeptides have not been stereospecifically assigned, since at ambient temperature the moderately fast flip around the N–C-bonds averages expected NOEs between the Hε-proton and the Hη21-protons. In the amino acid Arg the flip rate around the Nε–Cζ-bond is about 900 to 1000 s−1 at room temperature in the presence of 30% methanol-d6 (Henry and Sykes, 1995). As a consequence, the chemical shifts of the Hη-protons are averaged at 500 MHz at room temperature. At 263 K and at 500 MHz proton resonance frequency the rotation around the Nε–Cζ-bond is sufficiently slowed down in this solution for observing two separated Hη/Nη-cross peaks at (6.52, 70.78) ppm and at (6.97, 72.78) ppm. At 223 K in the presence of 50% methanol the rotation rate around the Cζ-Nη-bond is decreased in such a way that the two Hη signals bound to the downfield shifted nitrogen can be observed separately but not those of the highfield shifted nitrogen (Yamazaki et al. 1995). Unfortunately, stereospecific assignments have not been reported. In our tetrapeptide, the guanidino proton and nitrogen resonances are well separated at 283 K and at 800 MHz 1H-resonance frequency. A stereospecific NOE-based assignment cannot be performed at this temperature because of the motional averaging of the NOEs. However, the motions can be slowed down sufficiently by decreasing the pH to 2.4 and by decreasing the temperature to 260 K at 195 MPa where the solvent is still fluid. In the 3D-[1H, 15N]-NOESY-HSQC it shows a strong NOE from the Hε resonance at 7.29 ppm to one set of the Hη-resonances (data not shown). This indicates that the downfield shifted resonance at 6.98 ppm corresponds to Hη21/η22 bound to Nη2 under these experimental conditions. The assignment of the resonances at ambient conditions (Table 4) was performed by following the continuous temperature and pressure dependent shift changes. Interestingly, the Hη21/η22 resonance corresponds to the downfield shifted resonance that shows a smaller rotation rate around the Cζ–Nη-bond than the high field shifted resonance. This is in line with a small sterical hindrance by the Nε-group.
Imidazole nitrogen atoms of histidine
The histidine Nδ1 and Nε2 resonances were assigned by [1H,15N]-HSQC spectroscopy using the two-bond coupling to the ring protons. In agreement with this assignment are the chemical shift values given by Platzer et al. (2014) for Ac-GHG-NH2. In addition, a stronger pH dependence of chemical shifts is expected and observed for Nδ1.
Pressure dependence of 1H chemical shifts of side chain protons bound to a carbon atom
The resonances of side chain protons bound to a carbon atom are listed in Tables 2 and 3. Here, besides long range structural effects the pressure response is expected to depend on the type of the group the proton is attached to as well as on the position in the side chain.
Pressure dependent shifts of Hβ-protons
As an example, Fig. 1 shows the pressure response of the Hβ-resonances of histidine in the tetrapeptide that is clearly non-linear. Such a deviation from the linearity is observed for most of the other Hβ-resonances. The size and direction of the pressure induced shifts is strongly dependent on the amino acid under consideration. In Table 2 the parameters δ0, B1 and B2 obtained from a fit of the data are summarized. Most of the resonances show an upfield shift with pressure. In addition, the sign of the second order pressure coefficient B2 is opposite to that of the first order coefficient B1 for all values with a reasonable error estimate (Table 2). This leads to a saturation-like behavior where the pressure response gets weaker with higher pressures. In some residues, the two methylene resonances move in different directions with pressure, namely in the aromatic residues His, Phe, and Tyr as well as in Glu and cysteine (see e.g. Fig. 1). According to the SAIL data, the Hβ2-resonances are always shifted upfield relative to the Hβ3-resonances and have a negative first order coefficient B1. In contrast, the Hβ3-resonances have a B1 > 0, experiencing a downfield shift in the low pressure range. As a result, the two resonances become further separated with pressure. Since the absolute value of the first order pressure coefficient of the Hβ2-resonances is always larger than that of the Hβ3-resonances, the center of gravity of the two resonances moves upfield with pressure as also observed for all other unresolved methylene resonances except Asn and Asp. Here, both methylene resonances move downfield with pressure. This is probably due to the close-by carbonyl group of the side chain. In contrast to most of the methylene resonances, the resonances of the methine groups of Val and Thr shift downfield with pressure.
Pressure dependence of chemical shifts of the histidine Hβ- resonances in Ac-GGHA-NH2. Experimental conditions and fit parameters see Table 2, pH 8.5, temperature 283 K. (filled circle) Hβ2, (open circle) Hβ3 according to the tentative stereospecific assignment obtained by comparison with the SAIL amino acids
Pressure dependent shifts of side chain Hγ-, Hδ-, and Hε-methylene protons
The proton resonances of all γ-, δ-, and ε-methylene groups have a negative first order pressure coefficient (B1 < 0) and therefore shift upfield with pressure (Table 3). Most of them show a saturation-like behavior (B2/B1 < 0). Omitting all resonances with an uncertainty equal/larger the value itself (values in brackets in Figs. 2 and 3), significant exceptions are the Hγ2- and Hγ3-resonances of Glu and the Hγ13-resonance of Ile.
Pressure dependence of the guanidino resonances in Ac-GGRA-NH2. Experimental conditions and fit parameters see Table 4, pH 6.7, temperature 283 K, (open circle) Hη11/η12, (filled circle) Hη21/η22
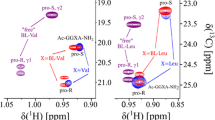
Influence of N- and C- terminal protection on chemical shifts and first order pressure coefficients in the model peptides. The chemical shifts δ0 and the first order pressure coefficients B1 of side chain protons of GGXA (Arnold et al.. 2002) at pH 5.4 und T = 305 K are plotted versus those of Ac-GGXA-NH2 at pH 6.7 and 283 K. a HN, b Hα, c Hβ, d HX with X other side chain protons. The linear correlation coefficients for δ0 for groups A to D are 0.90, 0.99, 0.99, and 0.99, respectively. For B1 they are 0.74, 0.65
Pressure response of methine protons in γ-position and methyl protons
The methine resonances of Leu and Thr shift again upfield with pressure (B1 < 0) as already described for the methine group of valine. Thr shows a clear saturation-like behavior since B2 is positive. For Leu the error of is so large that a positive or negative sign of B1 is allowed within the limits of error. The methyl resonances of Ile, Leu, Val, Met, and Thr, shift upfield with pressure and all of them exhibit a saturation like behavior (Table 3). An analogous pressure response is also observed for the methyl group of Ala in β-position (Table 2).
Pressure dependent shifts of protons in aromatic ring systems
All ring protons of Phe and Tyr show an upfield shift with pressure (B1 < 0) with a saturation like behavior (B2 > 0) (Table 3). This is also true for most of the ring proton resonances of Trp with exception of the Hδ1 and the Hζ2 resonances. The latter resonances have positive first and second order pressure coefficients and shift therefore downfield with pressure. Both resonances are relatively close to the ring nitrogen and may be influenced by the pressure dependent polarization of the NH group. In line with this hypothesis, also the proton resonances of His show an analogous pressure response at pH 4.0 and pH 8.5 with positive first order pressure coefficients. The B2/B1-ratio of the Hε1 –resonance of His at pH 4.0 is negative but positive at pH 8.5.
Pressure dependence of chemical shifts of side chain nitrogen and their directly bound hydrogen atoms
The resonances of side chain nitrogen atoms and their directly bonded hydrogen atoms are listed in Table 4. All side chain nitrogen resonances shift downfield with increasing pressure and show a slower increase of chemical shifts at higher pressures since B2 is always negative. The same behavior was observed earlier (Koehler et al, 2012) for the main chain nitrogen resonances.
Pressure response of the arginine guanidino group
The 1H as well as the 15N resonances of the arginine guanidino group are separated at 283 K and 800 MHz proton frequency and could be assigned stereospecifically (see above). At ambient pressure the Hη1 and Hη2 resonances are separated only by 0.44 ppm, the Nη1 and Nη2 resonances by 2.12 ppm (Table 4). At increasing pressure, the Hη1 resonances, that are highfield shifted relative to the Hη2 resonances, shift downfield and the Hη2 resonances shift upfield with pressure (Fig. 2). This means that these resonances become less separated at higher pressures. However, compared to their initial chemical shift difference this effect is rather small. Both Nη1 and Nη2 resonances first shift downfield with pressure (B1 > 0). At higher pressures they again become closer because of the more negative second order pressure coefficient of the downfield shifted resonance (Table 4). With higher pressure the corresponding proton resonances get broader probably because of the increased exchange rate with the water. Compared to the Nη resonances, the Nε resonance shows with 1.8 ppm GPa−1 a several times weaker pressure response that get smaller at high pressure. This is because the second order pressure coefficient has the opposite sign of the first order pressure coefficient. The corresponding proton resonance shifts upfield with pressure; the shift changes get stronger with higher pressures since here both pressure coefficients have the same negative sign.
Pressure response of resonances of the nitrogen and its attached protons of the histidine and tryptophan rings
The pressure dependence of the histidine nitrogen resonances could only followed at pH 4.0 with sufficient spectral quality. Under these conditions, the ring is positively charged and the two nitrogen atoms are protonated. The proton as well as the nitrogen resonances have positive B1-values that is, they shift downfield with pressure in our pressure range. Since the B2-values have signs opposite to the B1-values, at higher pressure the downfield shifts are attenuated. The B1- and B2-value of the Nε2 atom are approximately one order of magnitude larger than those of the Nδ1 that shows only a very small pressure dependence. At higher pH values, histidine exists in two tautomers, the τ- and the π-tautomer, with either the Nε2 or the Nδ1 being protonated. At pH 8.5 preferentially the Nε2 atom remains protonated and is therefore characterized by relatively small chemical shift changes with pH. In contrast, the Nδ1 atom shows much stronger pH-dependence of its chemical shift (Blomberg et al. 1977; Platzer et al. 2014) because of its deprotonation with increasing pH. This means that the relatively strong chemical shift response of the Nε2 atom is not due to a partial deprotonation because of a change of the pKa of the histidine ring, as this would primarily influence the Nδ1 shifts. The pressure response of the resonances of the protons directly bound to the ring nitrogens of histidine could not be estimated here since they are exchange broadened considerably.
The nitrogen resonance of Trp shows a chemical shift response similar to that of the histidine Nε2. It is characterized by a downfield shift with pressure that is slowed down at very high pressures. With a first order pressure coefficient of − 0.24 ppm GPa−1, an upfield shift with pressure of the attached proton is observed that is opposite in direction and smaller than the backbone amide shifts with an average first order pressure coefficient of 0.52 ppm GPa−1 (Koehler et al. 2012).
Pressure response of the lysyl side chain amino group
The nitrogen resonance of the lysyl amino group is shifting downfield with pressure as all side chain and backbone nitrogen resonances. The corresponding protons shift upfield with pressure, in contrast to the expectation that the a decrease of the N–H bond length with pressure would induce a downfield shift (Wagner et al. 1983; Asakawa et al. 1998; Li et al. 1998). The 1H linewidth of the amino group in the non-decoupled spectrum is approximately 62 Hz and is not influenced significantly by pressure at 283 K.
Pressure response of the amide group of Asn and Gln
The amide side chain nitrogen resonances of Asn and Gln show the largest pressure response of all side chain nitrogen atoms, with B1 values of 7.7 and 7.5 ppm GPa−1, respectively (Table 4). The observed downfield shifts with pressure are also significantly larger than the average backbone amide nitrogen shifts. Their mean B1 value is 2.91 ppm; the largest first order coefficient is found for Gly with 3.79 ppm GPa−1, still smaller than the side chain amide shifts (Koehler et al. 2012). The amide side chain protons of Asn and Gln were assigned stereospecifically earlier (Harsch et al. 2013). Both proton resonances shift downfield with pressure. The two protons show a different pressure response with a larger shift for Hδ22 in Asn as well as for Hε22 in Gln. In HPr from S. carnosus almost all (7 out of 9) Asn and Gln amide protons could be assigned stereospecifically by 3D-NOESY spectroscopy and their pressure response could be analysed (Kalbitzer et al. 2000). In all side chains the first order pressure coefficients of the two protons are positive as in the tetrapeptide. At 298 K, except of Asn38 the B1-values of the upfield shifted resonance (Hδ22 in Asn and Hε22 in Gln) are much larger than the downfield shifted resonances. However, at 278 K also in Asn38 the B1-value of Hδ22 is larger than that of Hδ21 indicating a temperature induced exchange averaging of the two values. Also the nitrogen first order pressure coefficients are relatively large and positive (average 7.67 ppm GPa−1 at 278 K). This value is very close to 7.6 ppm GPa−1 at 283 K, the mean value for Asn and Gln in our random-coil model although HPr is a quite rigid, well-folded protein. The hydrogen bonding expected in the protein appears to have no larger effect on the amide nitrogen pressure response. This is different for the hydrogen resonances: Here, the mean values for HPr at 278 K are with 0.22 and 1.11 ppm GPa−1 significantly larger than 0.12 and 0.40 ppm GPa−1 in the tetra peptide at 283 K indicating pressure induced changes in hydrogen bond lengths. The pressure coefficients of the amide side chains in the tetrapeptides predict that at lower pressures the shift difference between the two resonances decreases but increases again at pressures higher than 350 to 400 MPa, since the second order coefficients have different signs. It is expected that a similar behavior is found in the protein but no second order pressure coefficients have been determined here.
Influence of the N- and C-terminal protection on the pressure response
Protection of the N- and C-terminus of tetrapeptides by acetylation and amidation may influence also the pressure response of amino acid Xxx in position 3. The chemical shifts at ambient pressure δ0 and the first order pressure coefficients B1 of different groups of atoms of the unprotected tetrapeptides are plotted against the corresponding values in the protected tetrapeptides in Fig. 3. For the individual chemical shifts δ0 of the main chain amide protons significant deviations from the correlation line are observed, although the Pearson correlation coefficient is with 0.90 still quite high. For the other three groups of atoms investigated here (Hα, Hβ, HX) (Fig. 3) almost perfect correlations of the chemical shifts at ambient pressure are observed with correlation coefficients of 0.99 indicating that they are not influenced significantly by the protecting groups. The variations of the HN-shifts may mainly be due to the different experimental temperatures in the two data sets, since the temperature dependence of these shifts is known to vary from amino acid to amino acid (Jimenez et al. 1986; Kjaergaard et al.2011a). In general, for the first order pressure coefficients much larger deviations from a linear correlation are observed. A simple reason for these deviations may be that only the pressure response of the HN-resonances was fitted by a second degree polynomial by Arnold et al. (2002) but that of the other proton resonances only by a first degree polynomial. In contrast, in the present paper the data quality was high enough to fit all data with a second degree polynomial. In addition, temperature and pH is different in the two studies and protected tetrapeptides are compared with unprotected tetrapeptides where the termini are at least partly charged. In agreement with the effect of the fitting procedure, the correlation of the B1-values of the amide proton resonances is with 0.74 significantly higher than 0.65, 0.61, and 0.57 found for the Hα-, Hβ-, and the HX- (X, other side chain protons than Hα or Hβ) resonances, respectively (Fig. 3). A few amino acids show deviations of the first order pressure coefficients from the correlation line, the largest deviations are observed for the Hα of Asp and Glu, the Hβ and the Hζ of Lys (Fig. 3). For Asp and Glu pH-dependent differences in protonation states of the carboxyl groups may be the reason and/or the interaction of the charged side chain with the charged N-or C-termini in the unprotected tetrapeptides. For Hζ of Lys only the latter effect can apply. For GGEA the effect of the terminal charges on the pressure response was already published by Kremer et al. (2003), where the modification of the C-terminal carboxyl group strongly changes the pressure response. However, also polar uncharged residues such as Asn or Tyr show a strong effect on their pressure response when the terminal groups are modified. This again stresses the effects of the charged groups of the termini on the pressure response.
Neighborhood effects on the pressure response
The chemical shifts of a given amino acid in random-coil peptides are dependent on the next neighbors in the sequence. They can be corrected by using a simple additive model (Braun et al. 1994; Wishart et al. 1995a; Schwarzinger et al. 2001; Tamiola et al. 2010; Kjaergaard et al. 2011a; Kjaergaard and Poulsen 2011b). A similar effect from neighboring amino acids in the sequence can be expected for the pressure response. The data were evaluated analogously to the method applied by Schwarzinger et al. (2001) to the pentapeptide Ac-GGXGG-NH2 in 8 M urea at pH 2.3 and 293 K. However, since we have a tetrapeptide, only the correction factors \({C}_{\mathrm{1,2}}^{-1,a}\)(y), \({C}_{\mathrm{1,2}}^{+1,a}\)(y), and \({C}_{\mathrm{1,2}}^{+2,a}\)(y) for atoms a could be determined (see “Materials and methods” Section) with \({C}_{\mathrm{1,2}}^{-j,a}\)(y) correction factors for atom a in amino acid X in position i when amino acid y is located at position i + j in the sequence. The mean sequence correction factors calculated in our peptide for HN and Hα for the shifts at ambient pressure agree well with those obtained by the others groups. They are − 0.03, − 0.08, and 0.09 ppm for HN and − 0.03, − 0.03, − 0.03 ppm for Hα for Gly1, Gly2, and Ala4, respectively. In Ac-GGXGG-NH2, they are − 0.01, − 0.05, 0.15 ppm for HN and − 0.03 − 0.02, − 0.03 ppm for Hα for Gly1, Gly2, and Gly4 as reported by Schwarzinger et al. (2001). The average correction factors reported by Kjaergaard et al. (2011a) are even closer to our values with − 0.02, − 0.07, 0.09 ppm for HN and − 0.02, − -0.02, − 0.03 ppm for Hα for Gly1, Gly2, and Gly4. However, the individual values correlate relatively weakly between all three data sets. This is probably due to different experimental conditions, especially the urea concentration, the pH, and the experimental temperatures. Most of the correction factors for the first order pressure coefficients of amide backbone protons are positive (Table 5). The strongest effects on the pressure response are observed when the amino acid directly preceding amino acid X is not a Gly. In average, B1 is increased by 0.08 ppm GPa−1 and B2 decreased by 0.08 ppm GPa−2. Since also all experimental first order coefficients of amide protons are positive (Koehler et al. 2012), the positive correction factor leads to a larger downfield shift with pressure except of Ser, His and Gln that cause a quite small decrease of the pressure response. The largest corrections are observed for aromatic residues and amino acids with long side chains in position i−1. The maximum correction for B1 is required for Leu with 0.18 ppm GPa−1. The effect of amino acids following amino acid X is in the average smaller than that due to the preceding amino acid. It is similar in position i + 1 and i + 2. For the amino acid in position i + 1 the average corrections for the first order and second order pressure coefficient are 0.01 GPa−1 and − 0.05 ppm GPa−2; in position i + 2 they are 0.04 GPa−1 and − 0.05 ppm GPa−2.
The corrections for the Hα pressure induced shifts are substantially smaller than those for the amide protons. Again, the effects of the preceding amino acid are stronger than the effects of the succeeding amino acids. The average corrections required for B1 and B2 by amino acids in position i−1 are − 0.03 GPa−1 and 0.03 ppm GPa−2, respectively. The corresponding values for the succeeding amino acids are − 0.02 GPa−1 and 0.01 ppm GPa−2 (i + 1), and 0.00 GPa−1 and 0.01 ppm GPa−2 (i + 2).
Since most of the Hα resonances shift upfield with pressure (Beck Erlach et al. 2016), the negative correction factors for B1 values intensify the downfield shift. It has been observed earlier and also verified here for the side chain atoms that most second order coefficients have an opposite sign relative to the first order coefficient (Beck Erlach et al. 2016). The same is true for the correction factors, meaning that in most cases the curvature is enhanced by amino acids other than Gly in the neighborhood.
From our data set, also the correction factors \({C}_{\mathrm{1,2}}^{-1,a}\)(y) for the β-methyl group of Ala can be derived. The correction factors \({C}_{1}^{-1,a}\) vary between − 0.12 and 0.01 ppm GPa−1 and the correction factors \({C}_{2}^{-1,a}\) between − 0.05 and 0.03 ppm GPa−2 and thus are of the same order of magnitude than the corresponding B1 and B2 values themselves of − 0.022 ppm GPa−1 and 0.02 ppm GPa−2, respectively (Table 3 and 4).
Correlation between the second and first order pressure coefficients for different groups of side chain atoms
As shown earlier (Beck Erlach et al. 2014), under certain conditions the ratio of B2/B1 is related to the local compressibility. When the pressure response can be described by a two-state model with a free energy difference |ΔG/2RT|< < 1, B2/B1 equals—½ Δβ′/ΔV. Note that in the cited paper the definition of the second order pressure coefficient was different leading to a factor of two relative to B2 defined by Eq. 1 in the present paper. Δβ’ is the difference of the partial molar compressibility factors and ΔV the difference in the partial molar volumes. If this process describes the whole tetrapeptide, the obtained values should be identical for all atoms of a given tetrapeptide, if it describes a general feature of all tetrapeptides, e. g. the properties of the surrounding water shell, it should have the same value for all atoms and tetrapeptides within the limits of error. Figure 4 shows a plot B2 as a function of B1 for the Hβ- and the Hγ-resonances. Whereas the two quantities are relatively well correlated for Hβ-resonances with a correlation coefficient of − 0.82, the correlations for the Hγ-resonances and the Hδ-resonances are much smaller with correlation coefficients of − 0.48 and − 0.21, respectively, probably, since they represent a chemically more inhomogeneous group of atoms (Table 6). For the other side chain proton resonances the correlation is again larger, probably since these resonances form again a more homogenous group.
We have reevaluated the data partly presented by Beck Erlach et al. (2016, 2017) and Koehler et al. (2012). Relative high negative correlation ≤ − 0.82 are found for the side chain carbons (Table 6). Also quite high negative correlations were found for the main chain atoms HN, Hα, Cα, N, and C′ (Table 6, Beck Erlach et al. 2016) that form again chemically more homogeneous groups similar to the Hβ-resonances as already discussed. Ordering the resonances according to their chemical groups, e. g. methylene and methyl groups leads to similar correlation coefficients for B1 and B2 as found for ordering them according to their position in the side chain, indicating that assignment to a given chemical group represents a property independent of the position.
The slopes of the plot of B2 as function of B1 (see e. g. Fig. 4) corresponds to their ratios for different positions in the amino acids and different groups. They are listed in Tables 6 and 7.
For the side chain protons they vary in the range of − 0.18 (Hδ) and − 1.39 GPa−1 (Hβ), for the side chain carbons in the range of − 0.45 (Cζ) and − 0.90 GPa−1 (Cδ), for the main chain atoms between -1.07 (C′) and − 1.45 GPa−1 (HN) (Table 6). The values obtained from the slope are much more reliable than just the means calculated from the individual values in Tables 2 and 3 because the errors of the individual values are sometimes quite large. As an example, one would obtain mean values of B2/B1 of 0.4 ± 2 and − 0.31 ± 1.12 GPa−1 for the Hβ- and the Hγ-resonances (Tables 2 and 3), respectively, very different to the values given in Table 6. When considering distinct chemical groups, similar B2/B1-ratios are obtained. A significant exception is the pressure response of the nitrogen resonances of side chain amide groups where the B2/B1-ratio is positive with 1.39 GPa−1. Here, a specific pressure effect may become visible, e. g. the interaction of the NH2-group with its C=O-group.
Many B2/B1-ratios for the different groups and positions are the same within the limits of error indicating that at least partly a global two-site exchange may be involved in the observed pressure response.
Application of pressure coefficients of random-coil peptides
The simplest application of the random-coil pressure coefficients is the interpretation of pressure dependent chemical shifts of polypeptides. If the experimental shifts of a stretch of the sequence at ambient pressure are close to those predicted from the random-coil parameters, the probability is high that this region is disordered. This is even more likely when its pressure response approximates that of a random-coil model peptide as defined by its pressure coefficients. If this is true for any pressure than the probability is very high. It has been proposed earlier for peptides at ambient pressure, that also a secondary structure propensity can be derived from the chemical shift difference of the actual values from random-coil values (Yao et al. 1997). This procedure can now also be applied at data recorded at high pressure since the random-coil shifts at any pressure are now known. Such a prediction of the secondary structure propensity would allow a more informative interpretation of NMR spectra of intrinsically disordered proteins (see e.g. Roche et al. 2013).
Another important application of the model peptides is the thermodynamic evaluation of structural transitions of proteins (see e.g. Kalbitzer et al. 2013a; Kalbitzer 2015). When a protein exists in N structural states with M ≤ N states in fast exchange and the other states in slow or intermediate exchange, the pressure dependence of the chemical shift of a certain atom k (Baskaran et al. 2010) is given by
with pj the probability for state j; ΔP = P-P0 the difference of the actual pressure and the initial pressure (usually ambient pressure); \({\delta }_{j}^{k}(\Delta P)\) the chemical shift of atom k in state j as function of ΔP; ΔGij (ΔP) the difference between the free energy of state i and state j as function of ΔP; R the gas constant and T the absolute temperature. One has always to be aware that the terms slow, intermediate, and fast are measured relative to the NMR-time scale (essentially the chemical shift difference of the nucleus in different states) that may be different for any observed nucleus although only one global transition with a fixed absolute time scale is involved. In addition, note that the chemical shift \({\delta }_{j}^{k}\) generally is a function of pressure since the compression of the molecule per se causes a change of all chemical shift. The pressure dependence of ΔGij (Heremans and Smeller 1998) is given by
with \(\Delta {V}_{ij}^{0}\) the difference in the molar volume of state i and state j.
The function describing the change of \({\delta }_{j}^{k}\) by increasing pressure is not known and has to be approximated. It is caused by the anisotropic compression of the structure in state j combined with an amino acid specific chemical shift response. The functional dependence on pressure is not known a priori but the random coil data suggest that in general the pressure dependence of chemical shifts is non-linear and amino acid and atom specific. As in the case of random-coil peptides at least a second degree Taylor polynomial may be required (Eq. 1). A full fit of the compression effects on the chemical shifts \({\delta }_{j}^{k}\left(P\right)\) would then require three additional parameters per atom and conformational state to be fitted to the data together with three global parameters per transition required for the thermodynamic analysis, that is
It is obvious that the number of parameters is much too large to allow a stable fit of the data. With the assumption that the compression effects are (almost) identical in the different states, Eq. 7 can be simplified with the state independent pressure coefficients \({B}_{1}^{k}\) and \({B}_{2}^{k}\) to
A further simplification can be introduced by assuming that the pressure coefficients can be approximated by the corresponding random-coil values. This is equivalent to subtracting the function \({(B}_{1}^{k}\Delta P+ {B}_{2}^{k}{(\Delta P)}^{2}\) from the experimental chemical shifts before fitting the data as it is often done.
Conclusions
With this paper, together with the data previously published by Beck Erlach et al. (2016, 2017), we have a complete data set of the pressure response of a random-coil model peptides including all 1H-, 13C-, and 15N -resonances of the main and side chains. For the amide and Hα-resonances we now also provide a correction for different neighbors in the sequence.
A simple use of this data is to compare the pressure response of model peptides derived here with a protein whose structure is unknown. If the pressure response of the protein or part of the protein is equal to that derived for the random-coil peptides, it can be safely assumed that it is mainly disordered.
The other application is the use of these data for the description of the compression effects observed in a multistate protein as described above. Of course, these random-coil data cannot completely predict these effects since in general the compression effects are also influenced by the three-dimensional structure.
A third application of the high-pressure data is their application for developing theory. At the end only quantum chemical methods will be sufficient to calculate the pressure response of larger peptides. For the peptide bond model N-methyl-acetic acid (NMA) we could show that the pressure dependent chemical shift changes can be predicted rather well for the 1H, 13C, and 15N nuclei (Frach et al. 2016).
References
Akasaka K, Matsuki H (2015) High pressure bioscience: Basic concepts, applications and frontiers. Springer, Heidelberg
Arnold MR, Kremer W, Lüdemann HD, Kalbitzer HR (2002) 1H-NMR parameters of common amino acid residues measured in aqueous solutions of the linear tetrapeptides Gly-Gly-X-Ala at pressures between 0.1 and 200 MPa. Biophys Chem 96:129–140
Asakawa N, Kameda T, Kuroki S, Kurosu H, Ando S, Ando I, Shoji A (1998) Structural studies of hydrogen-bonded peptides and polypeptides by solid-state NMR. Ann Rep NMR Spectrosc 35:55–137
Baskaran K, Brunner K, Munte CE, Kalbitzer HR (2010) Mapping of protein structural ensembles by chemical shifts. J Biomol NMR 48:71–83
Beck Erlach M, Munte CE, Kremer W, Hartl R, Rochelt D, Niesner D, Kalbitzer HR (2010) Ceramic cells for high pressure NMR spectroscopy of proteins. J Magn Reson 204:196–199
Beck Erlach M, Koehler J, Moeser B, Horinek D, Kremer W, Kalbitzer HR (2014) Relationship between nonlinear pressure-induced chemical shift changes and thermodynamic parameters. J Phys Chem B 118:5681–5690
Beck Erlach M, Koehler J, Crusca E Jr, Kremer W, Munte CE, Kalbitzer HR (2016) Pressure dependence of backbone chemical shifts in the model peptides Ac-Gly-Gly-Xxx-Ala-NH2. J Biomol NMR 65:65–77
Beck Erlach M, Koehler J, Crusca E Jr, Munte CE, Kainosho M, Kremer W, Kalbitzer HR (2017) Pressure dependence of side chain 13C chemical shifts in model peptides Ac-Gly-Gly-Xxx-Ala-NH2. J Biomol NMR 69:53–67
Blomberg F, Maurer W, Rüterjans H (1977) Nuclear magnetic resonance investigation of nitrogen-15-labeled histidine in aqueous solution. J Am Chem Soc 99:8149–8159
Braun D, Wider G, Wüthrich K (1994) Sequence-corrected 15N “random coil” chemical shifts. J Am Chem Soc 116:8466–8469
Bundi A, Wüthrich K (1979) 1H-NMR parameters of the common amino acid residues measured in aqueous solutions of the linear tetrapeptides H-Gly-Gly-X-L-Ala-OH. Biopolymers 18:285–297
Frach R, Kibies P, Böttcher S, Pongratz T, Strohfeldt S, Kurrmann S, Koehler J, Hofmann M, Kremer W, Kalbitzer HR, Reiser O, Horinek D, Kast SM (2016) The chemical shift baseline for high-pressure NMR spectra of proteins. Angew Chem Int Ed 55:8757–8760; Angew Chem 128:8900–8904
Gronwald W, Kalbitzer HR (2004) Automated structure determination of proteins by NMR spectroscopy. Prog Nucl Magn Reson Spectrosc 44:33–96
Harsch T, Dasch C, Donaubauer H, Baskaran K, Kremer W, Kalbitzer HR (2013) Stereospecific assignment of the asparagine and glutamine side chain amide protons in random-coil peptides by combination of molecular dynamic simulations with relaxation matrix calculations. Appl Magn Reson 44:319–331
Harsch T, Schneider P, Kieninger B, Donaubauer H, Kalbitzer HR (2017) Stereospecific assignment of the asparagine and glutamine side chain amide protons in proteins from chemical shift analysis. J Biomol NMR 67:157–164
Henry GD, Sykes BD (1995) Determination of the rotational dynamics and pH dependence of the hydrogen exchange rates of the arginine guanidino group using NMR spectroscopy. J Biomol NMR 6:59–66
Heremans K, Smeller L (1998) Protein structure and dynamics at high pressure. Biochim Biophys Acta 1386:353–370
Jimenez MA, Nieto JL, Rico M, Santoro J, Herranz J, Bermejo FJ (1986) A Study of the NH NMR signals of Gly-Gly-X-Ala tetrapeptides in H2O at low temperature. J Mol Struct 143:435–438
Kainosho M, Torizawa T, Iwashita Y, Terauchi T, Ono AM, Güntert P (2006) Optimal isotope labelling for NMR protein structure determinations. Nature 440:52–57
Kainosho M, Güntert P (2009) SAIL-stereo-array isotope labeling. Q Rev Biophys 42:247–300
Kalbitzer HR, Görler A, Li H, Dubovskii PV, Hengstenberg W, Kowolik C, Yamada H, Akasaka K (2000) 15N and 1H NMR study of histidine containing protein (HPr) from Staphylococcus carnosus at high pressure. Prot Sci 9:693–703
Kalbitzer HR, Spoerner M, Ganser P, Hosza C, Kremer W (2009) Fundamental link between folding states and functional states of proteins. J Am Chem Soc 131:16714–16719
Kalbitzer HR, Rosnizeck IC, Munte CE, Puthenpurackal Narayanan S, Kropf V, Spoerner M (2013a) Intrinsic allosteric inhibition of signaling proteins by targeting rare interaction states detected by high-pressure NMR spectroscopy. Angew Chem Int Ed 52:14242–14246; Angew Chem 125:14492–14496
Kalbitzer HR, Spoerner M (2013b) State 1(T) Inhibitors of activated ras. In: Tamanoi F (ed) The enzymes. Academic Press, Burlington, pp 69–94
Kalbitzer HR (2015) High pressure NMR methods for characterizing functional substrates of proteins. In: Akasaka K, Matsuki H (eds) High pressure bioscience: basic concepts. Applications and frontiers, Springer, Heidelberg, pp 179–198
Kepner GR (2010) Saturation behavior: a general relationship described by a simple second order differential equation. Theor Biol Med Modell 7(11):1–13
Kitahara R, Hata K, Li H, Williamson MP, Akasaka K (2013) Pressure-induced chemical shifts as probes for conformational fluctuations in proteins. Prog Nucl Magn Reson Spectrosc 71:35–58
Kjaergaard M, Brander S, Poulsen FM (2011a) Random coil chemical shift for intrinsically disordered proteins: effects of temperature and pH. J Biomol NMR 49:139–149
Kjaergaard M, Poulsen FM (2011b) Sequence correction of random coil chemical shifts: correlation between neighbor correction factors and changes in the Ramachandran distribution. J Biomol NMR 50:157–165
Koehler J, Beck Erlach M, Crusca E, Kremer W, Munte CE, Kalbitzer HR (2012) Pressure dependence of 15N chemical shifts in model peptides Ac-Gly-Gly-X-Ala-NH2. Materials 5:1774–1786
Kremer W, Arnold MR, Brunner E, Schuler B, Jaenicke R, Kalbitzer HR (2003) High pressure NMR spectroscopy and its application to the cold shock protein TmCsp from the hyperthermophilic bacterium Thermotoga maritima. In: Winter R (ed) Advances in high pressure bioscience and biotechnology II. Springer, Berlin, pp 101–112
Li H, Yamada H, Akasaka K (1998) Effect of pressure on individual hydrogen bonds in proteins. Basic pancreatic trypsin inhibitor Biochemistry 37:1167–1173
Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wüthrich K (1998) Recommendations for the presentation of NMR structures of proteins and nucleic acids. Pure Appl Chem 70:117–142
Peterson RW, Wand AJ (2005) Self-contained high-pressure cell, apparatus, and procedure for the preparation of encapsulated proteins dissolved in low viscosity fluids for nuclear magnetic resonance spectroscopy. Rev Sci Instrum 76:094101
Platzer G, Okon M, McIntosh LP (2014) pH-dependent random coil 1H, 13C, and 15N chemical shifts of the ionizable amino acids: a guide for protein pKa measurements. J Biomol NMR 60:109–129
Raiford DS, Fisk CL, Becker ED (1979) Calibration of methanol and ethylene glycol nuclear magnetic resonance thermometers. Anal Chem 51:2050–2051
R Core Team (2019). R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna. URL https://www.R-project.org/
Roche J, Ying J, Maltsev AS, Bax A (2013) Impact of hydrostatic pressure on an intrinsically disordered protein: a high-pressure NMR Study of α-synuclein. ChemBioChem 14:1754–1761
Rosnizeck IC, Graf T, Spoerner M, Tränkle J, Filchtinski D, Herrmann C, Gremer L, Vetter IR, Wittinghofer A, König B, Kalbitzer HR (2010) Stabilizing a weak binding state for effectors in the human ras protein by cyclen complexes. Angew Chem Int Ed 49:3830–3833; Angew Chem 122:3918–3922
Rosnizeck IC, Spoerner M, Harsch T, Kreitner S, Filchtinski D, Herrmann C, Engel D, König B, Kalbitzer HR (2012) Metal-bis(2-picolyl)amine complexes as state 1(T) inhibitors of activated Ras protein. Angew Chem Int Ed 51:10647–10651; Angew Chem 124:10799–10804
Spoerner M, Karl M, Lopes P, Hoering M, Loeffel K, Nuehs A, Adelsberger J, Kremer W, Kalbitzer HR (2017) High pressure 31P NMR on guanine nucleotides. J Biomol NMR 66:141–157
Schwarzinger S, Kroon GJA, Foss TR, Chung J, Wright PE, Dyson HJ (2001) Sequence-dependent correction of random coil NMR chemical shifts. J Am Chem Soc 123:2970–2978
Tamiola K, Acar B, Mulder FAA (2010) Sequence-specific random coil chemical shifts of intrinsically disordered proteins. J Am Chem Soc 132:18000–18003
Wagner G, Pardi A, Wüthrich K (1983) Protein conformation and proton nuclear magnetic-resonance chemical shifts. J Am Chem Soc 105:5948–5949
Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD (1995a) 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J Biomol NMR 5:67–81
Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD (1995b) 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR 6:135–140
Yamazaki T, Pascal SM, Singer AU, Forman-Kay JD, Kay LE (1995) NMR Pulse schemes for the sequence-specific assignment of arginine guanidino 15N and chemical shifts in proteins. J Am Chem Soc 117:3556–3564
Yao J, Dyson HJ, Wright PE (1997) Chemical shift dispersion and secondary structure prediction in unfolded and partly folded proteins. FEBS Lett 419:285–289
Acknowledgements
Open Access funding provided by Projekt DEAL. This work has been supported by the DFG (FOR1979 and KA 647), the Humboldt Society, Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP-2010/12953-4), and the Human Frontier Science Program Organization (HFSPO).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Beck Erlach, M., Koehler, J., Munte, C.E. et al. Pressure dependence of side chain 1H and 15N-chemical shifts in the model peptides Ac-Gly-Gly-Xxx-Ala-NH2. J Biomol NMR 74, 381–399 (2020). https://doi.org/10.1007/s10858-020-00326-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-020-00326-w