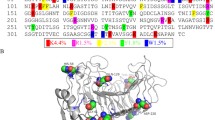
Abstract
The linker of the endoglucanase from Xanthomonas campestris pv. campestris ((PT)12) has a specific sequence, a repeating proline-threonine motif. In order to understand its role, it has been compared to a regular sequence linker, in this work—the cellobiohydrolase 2 from Trichoderma reesei (CBH2). Elastic properties of the two linkers have been estimated by calculating free energy profile along the linker length from an enhanced sampling molecular dynamics simulation. The (PT)12 exhibits more pronounced elastic behaviour than CBH2. The PT repeating motif results in a two-mode energy profile which could be very useful in the enzyme motions along the substrate during hydrolytic catalysis.






Similar content being viewed by others
References
Wriggers W, Chakravarty S, Jennings PA (2005) Control of protein functional dynamics by peptide linkers. Biopolymers 80(6):736–746
Beckham GT, Bomble YJ, Matthews JF, et al. (2010) The O-glycosylated linker from the Trichoderma reesei Family 7 cellulase is a flexible, disordered protein. Biophys J 99:3773–3781
Da Silva AC, Ferro JA, Reinach FC, et al. (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459–463
http://www.ncbi.nlm.nih.gov/protein/21232950?report=fasta. Accessed 19 June 2019
Shen H, Schmuck M, Pilz I, Gilkes NR, Kilburn DG, Miller R C Jr, Warren RA (1991) Deletion of the linker connecting the catalytic and cellulose-binding domains of endoglucanase A (CenA) of Cellulomonas fimi alters its conformation and catalytic activity. J Biol Chem 266(17):11335–11340
https://www.ncbi.nlm.nih.gov/protein/P07987.1?report=fasta. Accessed 19 June 2019
Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinform 9:40
Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5(4):725–738
MacKerell AD, Brooks CL, Nilsson L, Roux B, Won Y, Karplus M (1998) CHARMM: the energy function and its parameterization with an overview of the program (the encyclopedia of computational chemistry), vol 1. Wiley, Chichester, pp 271–277
Hui JP, White TC, Thibault P (2002) Identification of glycan structure and glycosylation sites in cellobiohydrolase II and endoglucanases I and II from Trichoderma reesei. Glycobiology 12:837–849
Sugita Y, Okamotor Y (1999) Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett 314:141–151
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802
Schneider T, Stoll E (1978) Molecular-dynamics study of a three-dimensional one-component model for distortive phase transitions. Phys Rev B 17:1302
Tuckerman ME, Martyna GJ, Berne BJ (1990) Molecular dynamics algorithm for condensed systems with multiple time scales. J Chem Phys 93:1287–1291
Anderson HC (1983) Rattle: “Velocity” version of the shake algorithm for molecular dynamics calculations. J Comput Phys 54:24–34
http://www.ks.uiuc.edu/Research/gpu/files/ICMS-Edinburgh/GB.pdf. Accessed 19 June 2019
Abraham MJ, Gready JE (2008) Ensuring mixing efficiency of replica-exchange molecular dynamics simulations. J Chem Theory Comput 4:1119–1128
Allen MP, Tildesley DJ (1987) Computer simulation of liquids. Oxford University Press, Oxford
Amadei A, Linssen AB, Berendsen HJ (1993) Essential dynamics of proteins. Proteins 17:412–425
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(1):33–38
Gomes TCF, Skaf MS (2012) Cellulose-builder: a toolkit for building crystalline structures of cellulose. J Comput Chem 33:1338–1346
Sammond DW, Payne CM, Brunecky R, Himmel ME, Crowley MF, Beckham GT (2012) Cellulase linkers are optimized based on domain type and function: insights from sequence analysis, biophysical measurements, and molecular simulation. Plos One 7:e48615
http://en.wikipedia.org/wiki/Proline. Accessed 19 June 2019
Romain C, Leonard S, de Brevern AG, Gelly JC (2014) PolyprOnline: polyproline helix II and secondary structure assignment database. Database: J Biol Databases Curation, 2009, 1–8
Rosseto FR, Manzine LR, de Oliveira Neto M, Polikarpov I (2016) Biophysical and biochemical studies of a major endoglucanase secreted by Xanthomonas campestris pv. campestris. Enzyme and Microb Technol 91:1–7
Villa E, Trabuco LG, Frank J, Schulten K (2008) Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16:673–683
Puhl AC, Prates ET, Rosseto FR, Manzine LR, Stankovic I, de Araújo SS, Alvarez TM, Squina FM, Skaf MS, Polikarpov I (2019) Crystallographic structure and molecular dynamics simulations of the major endoglucanase from Xanthomonas campestris pv. campestris shed light on its oligosaccharide products release pattern. Int J Biol Macromol 136:493–502. https://doi.org/10.1016/j.ijbiomac.2019.06.107
Prates É, Stankovic I, Silveira RL, Liberato MV, Henrique-Silva F, Pereira N Jr, Polikarpov I, Skaf MS (2013) X-ray structure and molecular dynamics simulations of endoglucanase 3 from Trichoderma harzianum structural organization and substrate recognition by endoglucanases that lack cellulose binding module. Plos One 8:e59069
Chichili VPR, Kumar V, Sivaraman J (2013) Linkers in the structural biology of protein-protein interactions. Protein Sci 22(2):153–167
George RA, Heringa J (2002) An analysis of protein domain linkers: their classification and role in protein folding. Protein Eng 15(11):871–879
Poon DKY, Withers SG, McIntosh LP (2007) Direct demonstration of the flexibility of the glycosylated proline-threonine linker in the Cellulomonas fimi xylanase Cex through NMR spectroscopic analysis. J Biol Chem 282(3):2091–2100
Funding
This study was financially supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) via grants 08/56255-9, 09/54035-4 and 10/08680-2; Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) via grants 490022/2009-0 and 550985/2010-7.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Skaf, M.S., Polikarpov, I. & Stanković, I.M. A linker of the proline-threonine repeating motif sequence is bimodal. J Mol Model 26, 178 (2020). https://doi.org/10.1007/s00894-020-04434-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-020-04434-0