Abstract
Coral reefs are undergoing degradation due to overexploitation, pollution, and climate change. Management and restoration efforts require that we gain a better understanding of the complex interactions between corals, their microbiomes, and their environment. For this purpose, Varadero Reef near Cartagena, Colombia, serves as an informative study system located at the entrance of the Bay of Cartagena adjacent to the Canal del Dique, which carries turbid and polluted water into the bay. Varadero’s survival under poor environmental conditions makes it a great study site for investigating the relationship between the microbiome and coral resistance to environmental stressors. To determine whether the microbiomes of Varadero corals differ from those in less impacted sites, we conducted a reciprocal transplant experiment by relocating coral fragments from Varadero as well as a geographically proximate reef that is less affected by plume dynamics (Rosario) across a gradient of turbidity (low, medium, and high). After 6 months of acclimatization, transplanted corals developed site-specific microbiomes that differed significantly from pre-transplant microbiomes, and corals transplanted to the highly impacted site from both Varadero and Rosario site saw higher mortality and an increase in overall microbial diversity. In combination with physiology and survivorship outcomes pointing to a limit in the corals’ photoacclimative capacity, our results indicate that, rather than surviving, Varadero Reef is experiencing a slow decline, and its corals are likely on the brink of dysbiosis. With continued anthropogenic interference in marine environments, sites such as Varadero will become increasingly common, and it is imperative that we understand how corals and their microbial symbionts are changing in response to these new environmental conditions.
Similar content being viewed by others
Introduction
Coral reefs worldwide are currently undergoing severe degradation, and anthropogenic disturbances have been the primary cause (Hoegh-Guldberg 2011; Hoegh-Guldberg et al. 2007). This is particularly true in the Caribbean, where a combination of overfishing, coastal pollution, and climate change have led to a decrease in coral cover of over 50% (Gardner et al. 2003; Hughes et al. 2003; Jackson et al. 2014; Mumby et al. 2007; Pandolfi et al. 2003). The degraded water quality conditions that result from human-derived disturbances can have severe effects on coral health, often increasing coral susceptibility to bleaching and disease (Kaczmarsky et al. 2005; Sutherland et al. 2010; Wagner et al. 2010; Wenger et al. 2016; Wooldridge 2009). While it is believed that most corals are unable to survive these changes, examples of reef “oases” that manage to escape severe declines in coral cover in otherwise degraded areas have begun to emerge across the globe (Guest et al. 2018). These include reefs surviving in sites with high sediment loads, believed to be unsuitable for coral growth due to lower light availability as well as increased fluctuations in temperature (Anthony and Larcombe 2000; Morgan et al. 2017; Van Woesik et al. 2012). Turbid, estuary-like reef environments have been created in other parts of the world (Browne et al. 2013; Fisher et al. 2019; Ziegler et al. 2019) due to human interference by way of shoreline pollution and development, such as dredging, that increase suspended sediment loads and subsequently reduce light availability, smothering corals in the process as well as reducing reproductive success and larval settlement (Anthony and Larcombe 2000; Bessell-Browne et al. 2017; Erftemeijer et al. 2012a, b; Shlesinger and Loya 2019). Reefs such as these have invited scrutiny by scientists aiming to better understand how and why some corals are able to survive under conditions so far removed from their natural habitats, and how these conditions affect susceptibility to disease and thermal stress.
Research on reef stress resistance requires a multifaceted approach that examines the coral holobiont as a whole (i.e., host, algal, viral, and microbial symbionts) and how each of its members responds to their environmental stressors (Bourne et al. 2016; Haas et al. 2016; Peixoto et al. 2017). We now know that the development and improvement of management and restoration efforts require that we gain a better understanding of the complex interactions between corals, their microbiomes, and their environment (Ainsworth et al. 2010; van Oppen and Blackall 2019; Peixoto et al. 2017; Sweet and Brown 2016).
A recent discovery by way of uniquely located reefs has been Varadero Reef, located at the entrance of Cartagena Bay, Colombian Caribbean. Varadero Reef extends over 1 km2 and is commonly thought to be a relic of what was once a dominant reef system in Cartagena Bay and nearby areas (López-Victoria et al. 2015; Pizarro et al. 2017). Currently, the reef maintains high coral cover and species diversity despite the high water column turbidity caused by freshwater discharges from the Magdalena river via the man-made distributary channel Canal del Dique (Pizarro et al. 2017; Restrepo et al. 2016, 2018). The major channeling works that occurred along the canal between 1951 and 1984 gradually transformed Cartagena Bay into a stratified estuarine system characterized by a surface layer of water with low salinity and high turbidity (Pizarro et al. 2017; Restrepo et al. 2017). The plume of salinity and turbidity extends out from the mouth of the channel and over Varadero Reef, exposing it to these conditions. Due to its sustained survival under poor environmental conditions for over a century (Vega et al. 2013), Varadero Reef has emerged as a promising study site for investigating the relationship between the coral microbiome and coral resistance to environmental stress, as well as microbial shifts in response to changes in water quality. To better understand the potential role of microbial members on the condition and survivorship of coral holobionts, we compared the microbial communities of corals from different sites with contrasting exposure to the perturbations caused by the Canal del Dique freshwater discharges and examined how these communities changed when corals were transplanted to new environments. We found that transplanted corals developed site-specific microbiomes that differed significantly from pre-transplant microbiomes, and corals transplanted to the highly impacted site (located at the mouth of Cartagena Bay) from both Varadero and Rosario sites saw higher mortality and an increase in overall microbial diversity.
Methods
Sample collection and experimental design
We conducted a reciprocal transplant experiment of corals between sites along a gradient of varying exposure to perturbations caused by the freshwater discharges from the Magdalena River via the Dique distributary channel. The Varadero site is located southwest of Cartagena Bay, nearly 6 km to the west of the Dique outlet (10° 18′ 23.3″ N, 75° 35′ 08.0″ W) (Fig. 1). The contrasting site used for this study, here referred to as Rosario Reef, has been less exposed to the canal-caused turbidity as it is located 21 km southwest of Varadero within the marine park, Parque Nacional Natural Corales de Rosario y San Bernardo (10° 11′ 12.1″ N, 75° 44′ 43.0″ W) and extends over 150 km2 (Díaz et al. 2000) (Fig. 1). A third site, here called Cartagena Bay, was used as a reference point for a more extreme version of the environmental perturbation that Varadero corals are experiencing (10° 18′ 04.5″ N, 75° 34′ 38.5″ W). This site is located at the inner side of the mouth of the Bay and contains no corals likely due to the increased turbidity and low-light conditions (though scattered and severely eroded coral exoskeletons are present). Preliminary analyses of the diffuse attenuation coefficient for downwelling irradiance (Kd) were obtained by measuring light intensities at different depths using the cosine-corrected PAR sensor of a Diving PAM (Walz, Germany). The Kd’s corroborated the contrasting optical properties of water column at each site (Table 1) and indicated that the diurnal light exposure at 3.5 m in Varadero, 3 m in Cartagena Bay, and 12 m in Rosario was similar. Irradiance was monitored during the study period with cosine-corrected light sensors (Odyssey submersible PAR logger, Dataflow systems, New Zealand), which were previously calibrated against a factory-calibrated LI-1400 PAR sensor (LI-COR, USA). The light sensors were cleaned and downloaded periodically to avoid biofouling, discarding data with evident cumulative light signal loss. The daily integrated irradiance indicated that all sites were consistent with low-light environments (Anthony and Hoegh-Guldberg 2003; DiPerna et al. 2018) (Table 1). The temperature was recorded every 30 m with HOBO pendant dataloggers (UA-002-64, Onset Computer Corporation, USA), and it was found to be similar among sites (Table 1).
Study area showing the three transplant sites with contrasting exposure to the canal-caused turbidity. Cartagena Bay was the closest site to the mouth of Canal del Dique exposed to the highest turbidity, followed by Varadero, and lastly Rosario. The diurnal light exposure at the chosen depths on each site (3, 3.5 and 12 m, respectively) was expected to be similar due to the optical properties of the water column resulting from the Dique plume influence. Only Rosario and Varadero served as donor sites
The coral Orbicella faveolata was chosen as an indicator species of the effects of suboptimal environmental conditions on reef-corals microbiome and physiology due to its importance as a major reef builder in the Caribbean. On October of 2016, flat fragments (~ 10 cm2) were collected from the edge of 15 coral colonies (n = 45 per site) at the two donor sites (Varadero and Rosario). Coral colonies were chosen randomly at a constant depth of ~ 3.5 m and ~ 12 m in Varadero and Rosario, respectively. Fragments were fixed with non-toxic epoxy (Z-Spar A-788 epoxy) to PVC panels placed at the same depth as donor colonies from each site. After acclimation to the local environment (2 weeks), corals were transplanted from their naturally turbid (Varadero) and clear (Rosario) environment in equal proportions (n = 15) to each of the three contrasting sites. Before and 7 months after transplantation (May of 2017), a series of physiological and genetic analyses were performed on corals.
Fragment sample collection was performed using hammers and chisels to remove small fragments from targeted corals. Divers wore gloves at all times to prevent contamination. Fragments were placed in Whirlpack bags and immediately flash frozen once brought to the surface. Tissue was separated from the coral skeleton using pressurized air, and the resulting tissue slurry was stored at − 80 °C until DNA extraction. Water microbiome sample collection was performed by passing 1 L of water through a 0.2 micron Sterivex™ filter. Filters were wrapped in sterilized aluminum foil and flash frozen, then stored at − 80 °C until DNA extraction.
DNA extraction and 16S sequencing
Fifty µL of DNA was extracted from all pre-transplant, mother colony, and surviving post-transplant samples using the MoBio Powersoil DNA Isolation Kit (MoBio Laboratories, Carlsbad, California). We performed two-stage amplicon PCR on the V4 region of the 16S small subunit prokaryotic rRNA marker gene, a region that is commonly used to target bacterial and archaeal communities. We used modified versions of the 515F (5′-ACA CTG ACG ACA TGG TTC TAC AGT GCC AGC MGC CGC GGT AA-3′) and 806R (5′-TAC GGT AGC AGA GAC TTG GTC TGG ACT ACH VGG GTW TCT AAT-3′) primers that include common sequence 1 (CS1) and common sequence 2 (CS2) linkers at the 5′ end, the universal primer sequences that are required for Illumina MiSeq amplicon tagging and indexing (Apprill et al. 2015; Caporaso et al. 2012). PCR reactions were prepared with 9 µL 5Prime HotMaster Mix (VWR International), 1 µL forward primer (10 µM), 1 µL reverse primer (10 µM), 1 µL of template DNA, and 8 µL of PCR-grade water. The PCR amplification was structured as follows: 3 min of denaturation at 94 °C; 30 cycles of 45 s at 94 °C, 60 s at 50 °C, and 90 s at 72 °C; then finally 10 min at 72 °C. Once the PCR was finished, samples were held at 4 °C before sequencing.
Amplicons were barcoded with Fluidigm Illumina primers and pooled for sequencing. The amplicon pool was then purified with AMPure XP beads and sequenced on the Illumina MiSeq sequencing platform at the DNA Services Facility at the University of Illinois at Chicago. Initial pre-processing of 16S sequences was performed using the Quantitative Insights Into Microbial Ecology package (QIIME; v1.9) (Caporaso et al. 2010). Paired end reads were joined and then demultiplexed. Chimeric sequences were identified and removed using USEARCH 6.1 as well as the QIIME default reference file. OTUs were picked at 97% similarity using USEARCH 6.1 and the pick_open_reference_otus.py command. The maximum amount of accepts and rejects were set to 16 and 64, respectively, and sized-based preference in clustering was enabled. Taxonomy was assigned using UCLUST. The resulting BIOM file was imported into R (v. 3.6.0) for downstream analysis and figure generation using the phyloseq, vegan, microbiome, DESeq2, and ggplot2 packages (Anders and Huber 2010; Ginestet 2011; Lahti et al. 2017; McMurdie and Holmes 2013; Oksanen et al. 2007).
16S analysis
Bray–Curtis, weighted UniFrac, and unweighted UniFrac dissimilarity matrices were constructed using OTU-level abundances. Significant differences in bacterial communities between sample types were assessed by PERMANOVA with Bray–Curtis and UniFrac distances and explanatory variables including transplantation status, site of origin, season, and depth using the adonis function from the vegan package. Models that encompassed both species were tested, as well as species-specific models. PCoA ordination was conducted to visualize the gradients of variation in the microbial communities across different environmental and physical variables. Alpha diversity values (Chao1) were determined for surviving post-transplant fragments to determine differences in richness between sites. A LefSe analysis was also conducted to identify environmentally explicit taxa (Segata et al. 2011).
Coral photo-physiology
Photosynthetic parameters of corals were obtained from PE (photosynthesis vs. irradiance) curves conducted under laboratory conditions. A custom-made water-jacket acrylic chamber with four independent hermetic compartments (~ 650 mL each) was used to run the PE curves, maintaining a constant temperature of 28 °C, close to natural conditions, with an external circulating water bath (Isotemp, Fisher Scientific). During each incubation, corals were submerged in filtered seawater (0.45 µm) under constant agitation by magnetic stirrers. Ten levels of irradiance between 0 and ~ 1400 µmol quanta m−2 s−1 were supplied at 10-min intervals with four 26 W LED bulbs (UL PAR38, LED Wholesalers Inc, USA). The light intensity was controlled with a custom-made software. The LEDs were operated in continuous mode with a multifunction I/O card (USB-6001, National Instruments Corp., USA) to avoid potential artefacts related to the effect of different pulsating frequencies on photosynthesis (Iluz et al. 2012). Oxygen concentrations inside the compartments within the chamber were measured with a 4-channel fiber optical oxygen meter system (FireSting, Pyroscience, Germany). The photosynthetic efficiency (α), compensating irradiance (Ec), saturating irradiance (Ek), respiration rates (Rd), and maximum photosynthetic rates (Pmax), were calculated following Iglesias-Prieto and Trench (1994) and Osinga et al. (2012).
Areal chlorophyll a (Chl a) content per unit of coral surface area was determined after obtaining coral tissue slurries with the help of an air gun connected to a scuba tank. Pigment extraction was performed in acetone/dimethyl sulfoxide (95:5 vol/vol) after homogenizing the slurries with a Tissue-Tearor Homogenizer (BioSpec Inc, USA). Chl a density was estimated spectrophotometrically with a modular spectrometer (Flame-T-UV–VIS, Ocean Optics Inc., USA) following the equations of Jeffrey and Humphrey (1975).
Results
Transplant survivorship
Fragment survivorship was determined in May of 2017, 6 months from the original transplant date in October of 2016 (Supplementary Fig. 1). All fragments that were transplanted from Rosario to Varadero (n = 15) and from Varadero to Varadero (self-transplanted controls, n = 20) survived. Survivorship varied both in Rosario and in Cartagena. In Rosario, Varadero fragments suffered 40% mortality (n = 15 transplanted) and self-transplanted Rosario controls suffered 20% mortality (n = 15) (Supplementary Fig. 1). In Cartagena however, fragments that originated from Varadero (n = 15) had higher survivorship than fragments from Rosario (n = 15), with only 27% mortality vs. 53%, respectively (Supplementary Fig. 1). No significant differences in mortality were found between fragments based on origin site or transplant site (p = 0.148; Kruskal–Wallis rank-sum test).
Pre-transplant and post-transplant microbial communities
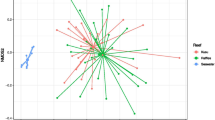
Post-processing and after removing samples with less than 1000 reads as well as removing singletons, the complete data set included 5,373,432 reads corresponding to 39,399 OTUs and 183 samples, including 43 Rosario pre-transplant fragments, 45 Varadero pre-transplant fragments, 14 and 13 Rosario and Varadero mother colonies, respectively, and 68 post-transplant fragments. Only surviving post-transplant fragments were sampled for 16S sequencing. The average number of OTUs per sample was 20,206; however, both fragments that were transplanted to Varadero and Cartagena Bay had higher average OTU numbers (11,043 and 11,230, respectively) than fragments transplanted to Rosario (7737). Furthermore, average OTU counts for pre-transplant fragments and mother colony samples were higher than those of post-transplant samples (ranging from 21,635 to 30,630). Seven water samples (n = 3 Rosario, n = 3 Varadero, and n = 1 Cartagena Bay) were collected during the second time point and were included in the supplementary material for comparison (Supplementary Fig. 2) but not in the main analysis given the low sample size. Overall, water microbiomes across all sites were distinct from all coral tissue samples, regardless of site and time point (Supplementary Fig. 2). Microbial communities across all samples (pre-transplant, post-transplant, and mother colonies) were found to be significantly structured by transplantation status, site of origin, status (hereby referring to the combination of origin site and transplantation site, i.e., specific transplantation treatment), and depth (p = 0.001, p = 0.001, p = 0.001, and p = 0.049, respectively; PERMANOVA on Bray–Curtis; Supplementary Table 1). Varadero mother colonies and Rosario mother colonies differed significantly in their microbial communities, as did Varadero pre-transplant fragments and Rosario pre-transplant fragments (p = 0.012 and p = 0.003, respectively; pairwise PERMANOVA on Bray–Curtis). A PCoA revealed strong clustering patterns by time point, with post-transplantation samples clustering significantly tighter than pre-transplant and mother colony samples (Fig. 2).
Principal coordinates analysis (PCoA) of Bray–Curtis matrices and family level bar plots of the microbial communities associated with samples from different transplant time points and treatments. a Clustering patterns across all sample groups, pre- and post-transplantation; b only post-transplant fragments, clustering by transplantation site regardless of origin; c family level bar plot of the top 12 most abundant OTUs in post-transplant fragments, calculated by site to which fragments were transplanted. “Low abundance” category contains all OTUs not in the top 12
While the clustering in Fig. 2a suggests little to no significant differences between all post-transplantation samples, post hoc analyses indicated that all post-transplantation sample types were significantly different from each other, with the exception of Varadero to Varadero fragments vs. Rosario to Varadero fragments as well as Varadero to Cartagena Bay fragments vs. Rosario to Cartagena Bay fragments (p = 0.1828 and p = 0.1338, respectively; pairwise PERMANOVA on Bray–Curtis; Supplementary Table 2). Therefore, while the PCoA plot suggests a convergence toward similar microbiomes regardless of origin and final transplant site, the statistical analyses point toward the existence of fine-scale yet significant differences between groups mainly influenced by transplant location rather than origin of fragment (except in the case of fragments moved to Rosario). This is reflected in Fig. 2b, as post-transplant fragments cluster based on transplant location (Fig. 2).
Family level abundances corroborate clustering patterns, as sample microbiomes have similarly abundant members depending on the site to which they were transplanted, regardless of origin site (Fig. 2c). Rosario fragments were found to have higher abundances of OTUs belonging to the Alteromonadaceae family compared to Varadero and Cartagena Bay fragments. In turn, Varadero and Cartagena Bay fragments had higher abundances of OTUs belonging to the Saprospiraceae family and the class Phycisphaerae (order CM11a). Cartagena Bay samples in particular had a higher abundance of Verrucomicrobiaceae and Phycisphaerae than both Varadero and Rosario samples. Clustering patterns of between post-transplant fragments and pre-transplant fragments were also reflected in OTU abundances, as pre-transplant fragments from both Rosario and Varadero contained more OTUs from the families Alphaprroteobacteria and Pirellulaceae (Supplementary Fig. 3). Pre-transplant fragments also had a greater number of OTUs belonging to the Planctomycetes phylum (Supplementary Fig. 3).
Differentially abundant taxa across post-transplant sites
To identify environmentally explicit taxa across transplant sites, we compared the microbiomes of all post-transplant fragments across sites using LEfSe (Fig. 3). LDA scores indicate that Cartagena Bay samples are enriched for Actinobacteria, taxa in the order Vibrionales, and Pseudomonadales. In turn, Varadero fragments were characterized by an enrichment of Proteobacteria and Rhizobiales, and Rosario fragments were enriched for groups including Alteromonadales and Idiomarina. The most differentially abundant taxon across all sample groups was Proteobacteria in Varadero fragments (LDA score (log 10) > 5). Overall, Cartagena Bay fragments were significantly enriched for 56 OTUs (compared to 7 in Varadero and 20 in Rosario), indicating greater bacterial diversity in these fragments. Alpha diversity analyses using Chao1 values confirmed these results, with Cartagena Bay samples having significantly higher OTU richness than Varadero or Rosario fragments (Supplementary Fig. 4, Wilcoxon rank-sum test, p < 0.0001). No significant difference in richness was found between Varadero fragments and Rosario fragments.
LEfSe analysis along with LDA scores of coral microbiota (relative abundance) in post-transplant fragments from Cartagena Bay, Rosario, and Varadero, respectively (p < 0.001, Kruskal–Wallis test). OTUs belonging to the same taxonomical group and with the same LDA score have been consolidated under one bar, with numbers added to the right of the bar indicating the number of OTUs being represented by that group. Candidate bacteria with uninformative taxonomy information (i.e., class A712011) were consolidated under a higher-level taxonomy for legibility and ease of identification
Coral photo-physiology
Most physiological parameters did not show a significant difference between the two source sites (Rosario at 12 m and Varadero at 3.5 m) before the transplant experiment, indicating the occurrence of similar phenotypes as can be expected due to the similar diurnal light exposure inferred from the Kd analysis prior to the sites’ selection. Only the specific absorption coefficient of Chl a\(\left( {{\text{a}}_{{{\text{Chl}}\,a}}^{*} } \right)\) was significantly lower in corals from Varadero (0.020 ± 0.002 m2 mg Chl a−1) relative to Rosario (0.041 ± 0.016 m2 mg Chl a−1) (t(7) = 3.83, p = 0.006). Chlorophyll a content per surface area was higher in corals from Varadero (109.33 ± 26.96 mg Chl a m−2) compared to Rosario (91.36 ± 44.15 mg Chl a m−2) but not significantly (supplementary material).
Seven months after transplantation, no significant differences were found in any physiological parameters between corals from Rosario and Varadero at each transplant site, suggesting similar photoacclimation capacity to local conditions (supplementary material). Significant differences in photosynthetic parameters, particularly in Ec (compensation point) and Rd (dark respiration rate) (Fig. 4), suggest the occurrence of photoacclimative responses of corals to lower irradiances in Cartagena Bay among the three sites. The occurrence of these photoacclimation responses is consistent with the lowest values of daily integrated irradiance recorded at this site (Table 1). The chlorophyll a content per surface area and the specific absorption coefficient of Chl a\(\left( {{\text{a}}_{{{\text{Chl}}\,a}}^{*} } \right)\), which covaries as a function of changes in chlorophyll a concentration, were similar between corals from Varadero and Cartagena Bay, but significantly different to corals from Rosario (Fig. 4).
Physiological responses of Orbicella faveolata to the transplant experiment between sites with contrasting exposure to the perturbations caused by the Dique plume. Cartagena Bay is the site most exposed to the turbid plume (orange bars), followed by Varadero (brown bars), and lastly Rosario which is significantly less exposed (blue bars). Change of parameters is relative to the averaged values obtained pre-transplantation. Different letters indicate significant differences (Tukey’s test, p < 0.05)
Discussion
The steep decline of coral reefs today is due in no little part to degraded water quality conditions caused by human activities (De’ath and Fabricius 2010; Kline et al. 2006; Vega Thurber et al. 2014; Wenger et al. 2016). As a result, it is becoming increasingly important to study the coral holobiont’s response to impacted conditions. The coral microbiome in particular has been shown to be highly sensitive to environmental stressors as well as to play a key role in coral health and survivorship (van Oppen and Blackall 2019; Zaneveld et al. 2016; Ziegler et al. 2017, 2019). Studies have shown that coral hosts, and subsequently their microbiomes, can have differential responses to environmental stressors depending on a number of different factors, including host species and location (Klaus et al. 2007; Lee et al. 2015; Röthig et al. 2016; Zaneveld et al. 2017). To help untangle the relationship between the microbiome and coral host survivorship and physiological responses to anthropogenically derived stressors, we conducted a reciprocal transplantation experiment with O. faveolata corals between three sites representing a gradient of turbidity impact off the coast of Cartagena, Colombia. Coral fragments from the low-impact site Rosario and the medium-impact site Varadero were assessed for both their physiological and microbial community level responses to different levels of turbidity.
Varadero and Rosario fragments, alongside mother colonies, differed significantly in their microbial communities before transplant. After 6 months of transplantation, we saw a convergence of fragment microbial communities toward microbiome states/“footprints” unique to each transplant site. Here, we use the term footprint to refer to microbial assemblages that are unique to each site and characteristic of fragments transplanted to those sites. Moreover, fragments moved to Rosario harbored significantly different microbial communities based on origin site, though these communities still differed from those of fragments moved to Cartagena Bay and Varadero. Studies that have conducted similar transplant experiments across gradients of stress (such as turbidity and temperature) have observed similar site-specific shifts in certain coral species, including Acropora hyacinthus and Acropora hemprichii in the Indo-Pacific (Ziegler et al. 2017, 2019). While other reciprocal transplant studies have observed sample microbiomes returning to normal pre-transplant states when moved back to their native site (Ziegler et al. 2017, 2019), our self-transplanted controls did not return to pre-transplant mircobiome states and did not resemble either the pre-transplant fragment microbiomes or the mother colony microbiomes. This could be due to a dramatic change in size, from colony-sized microbial community structures and assemblages to fragment-size dynamics, as a study by Pollock et al. (2018) has shown that microbial community composition can be affected significantly by colony size (Pollock et al. 2018). Similar results were also found by Williams et al. though their study also included age as a determining factor for the coral microbiome (Williams et al. 2015). Our study did not take mother colony size into account when selecting colonies for the transplant, which may account for the comparatively higher variability in the mother colony sample group. However, the influence of age on microbiome composition would seem to be trumped by the influence of colony size given our results, as all post-transplant samples showed low variability regardless of the original age/size of their mother colony. Studies have also shown that Orbicella spp. can be particularly sensitive to stress events, with immune system suppression having been shown to last up to a year after a bleaching event (Pinzón et al. 2015). While some species of coral have been shown to recover their bacterial communities in a matter of days following a disturbance, as found by Sweet et al. after treating Acropora muricata fragments with antibiotics, research shows that in the face of environmental disturbances microbiomes will be more plastic or less likely to recover depending on the species of their host as well as on the type of disturbance itself (Sweet et al. 2012). It is therefore possible that the act of transplantation itself may have had long-term effects on the coral fragments’ microbiomes, from which the fragments had yet to recover after 6 months.
Dramatic shifts toward different microbial states across sites indicate high microbiome flexibility and sensitivity to changes in turbidity levels in O. faveolata corals of this region. Interestingly, Orbicella corals have also been known to be promiscuous with their algal partners (Symbiodiniaceae), often harboring multiple genera at a time (DeSalvo et al. 2010; Kemp et al. 2014; LaJeunesse 2002; Lajeunesse and Trench 2000; Lewis et al. 2019) as well as exhibiting high immune plasticity and long-term stress effects (Pinzón et al. 2015). The observed shifts in the fragments’ microbial communities may either be the result of adaptive responses to their environment, or be directly caused by environmental factors such as poor water quality/excess opportunistic bacteria in the more turbid environments. Our data provide evidence toward the latter for a number of reasons.
First, there is the significantly increased richness/diversity in Cartagena Bay samples, as well as the greater number of differentially abundant taxa in those samples. Increases in microbial community diversity are common in stressed corals (Jessen et al. 2013; Morrow et al. 2012; Zaneveld et al. 2016), and could potentially be due to the coral host’s inability to regulate its microbiome when stressed, a theory outlined by Zaneveld et al. (2017) in their application of the Anna Karenina principle to animal microbiomes (Zaneveld et al. 2017). Further, physiology results indicate that while corals in our study exhibited photoacclimative responses to the lower light conditions of Cartagena Bay, the higher mortality of transplant fragments at the Cartagena site in conjunction with increasing mortality of coral colonies at depths lower than 10 m at the Varadero site point to a limit to the corals’ photoacclimatization capacity. The Cartagena Bay site is also more directly exposed to the turbid plume as well as high sedimentation, increasing the number of stressors and terrestrial bacterial sources acting on the fragments together with a potential energetic imbalance derived from acute light-limiting conditions. For example although Actinobacteria and Actinomycetaceae can be aquatic, these groups are representative of bacteria that are important in soil systems and have been found in increased abundance in Caribbean Acroporid corals due to nutrient runoff (Shaver et al. 2017). A particular species of soil bacterium known to be pathogenic to plants, Pseudomonas viridiflava, was also found to be significantly associated with Cartagena Bay samples, further evidence the terrestrial sources affecting this particular site (Sarris et al. 2012). An investigation of potential aquatic and coral-specific pathogens led us to the phylum SBR1093, commonly found in contaminated water and soil, which was also found to be significantly associated with Cartagena Bay samples (Wang et al. 2014). Also in Cartagena Bay samples were OTUs belonging to Pseudomonadales, Vibrionales, and Helicobacteraceae, which have been found in association with coral diseases including White Plague, Black Band, and White Syndrome (Ben-Haim et al. 2003; Kushmaro et al. 2001; Sunagawa et al. 2009; Sweet and Bythell 2015). The bacteria have also been found in unprotected reef sites with high human activity (Bruce et al. 2012). The microbial composition of the Cartagena Bay samples appears to be representative of their environment and suggestive of their overall health, explaining the higher mortality at this site.
In contrast to Cartagena Bay, the Varadero site exhibits an attenuation effect due to the more indirect exposure, leading to increased light availability, less sediment-induced stress, and occasional shading by the plume. Studies have shown that increased turbidity can occasionally act in ways that are beneficial to corals by protecting the affected reefs from increased temperatures as well as providing the opportunity for heterotrophy, both of which may explain the distinct lack of fragment mortality at Varadero (Morgan et al. 2017; Van Woesik et al. 2012). Microbes associated with Varadero corals included Bradyrhizobiaceae, a group of bacteria known to be involved in nitrogen cycling in terrestrial environments (Damjanovic et al. 2020), as well as Actinobacteria, which may engage in antimicrobial activity (Mahmoud and Kalendar 2016). Alphaproteobacteria are known to be widely associated with coral reefs and healthy coral tissue but may be replaced with other bacterial types when conditions become adversary (Lima et al. 2020). Interestingly, members of the alphaproteobacteria are also known to contain viral-like particles called gene transfer agents (GTAs) (McDaniel et al. 2010). GTAs are thought to randomly transfer genomic fragments widely among different bacterial genera, potentially providing a mechanism for rapid acquisition of beneficial traits on coral reefs (McDaniel et al. 2010).
Further analysis is required to be able to determine how shifts in the microbiome are affecting coral holobiont health and function. This would involve an in-depth categorization of the bacterial taxa found to increase/decrease significantly in response to these different turbidity gradients, as well as a functional analysis of these communities to determine whether they are acting in a way that is harmful (i.e., potentially pathogenic, opportunistic, or in other ways stressing) or beneficial to the coral host. A functional analysis is particularly important given the possibility that certain bacteria/archaea normally living in healthy symbiosis with the coral can carry pathogenic genes that may be activated by changes in their environment (Lesser et al. 2007; Rutherford and Bassler 2012). While it is likely that the shifts in these taxa are affecting holobiont metabolic pathways and cycling, not enough is known about coral-associated bacteria and their role in these processes to be able to determine how shifts in the groups we have identified are affecting coral health.
As climate change continues to devastate marine environments, it is becoming increasingly important to determine whether corals such as those found in Varadero are beginning to adapt to these changes in a way that is positive and can confer resistance and increased survivorship in the face of other stressors. While Varadero Reef’s history of survival under the stress of turbidity suggested a level of resistance, both physiological and microbiome data point to Varadero Reef’s slow albeit steady decline. Given the noted reduction in coral cover starting at a depth of ~ 10 m and continuing until 12–14 m at which point the low-light conditions no longer permit coral growth, there seems to be a gradual confinement of Varadero corals to shallow areas. As a result, it is likely that Varadero corals are being pushed toward the edge of dysbiosis, and we are now observing them on that precipice. The addition of further stress events, such as the spikes in temperature that have become increasingly common in the past couple of years, could therefore be the catalysts to the disappearance of this reef. With continued anthropogenic interference in marine environments, sites such as Varadero will become increasingly common, and it is imperative that we understand how corals and their microbial symbionts are changing in response to these new environmental conditions.
References
Ainsworth TD, Thurber RV, Gates RD (2010) The future of coral reefs: a microbial perspective. Trends Ecol Evol 25:233–240
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106
Anthony KRN, Hoegh-Guldberg O (2003) Variation in coral photosynthesis, respiration and growth characteristics in contrasting light microhabitats: an analogue to plants in forest gaps and understoreys? Funct Ecol 17:246–259
Anthony KRN, Larcombe P (2000) Coral reefs in turbid waters: sediment-induced stresses in corals and likely mechanisms of adaptation. In: Moosa MK et al Proceedings of the Ninth International Coral Reef Symposium, pp 239–244
Apprill A, McNally S, Parsons R, Weber L (2015) Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75:129–137
Ben-Haim Y, Thompson FL, Thompson CC, Cnockaert MC, Hoste B, Swings J, Rosenberg E (2003) Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int J Syst Evol Microbiol 53:309–315
Bessell-Browne P, Negri AP, Fisher R, Clode PL, Jones R (2017) Impacts of light limitation on corals and crustose coralline algae. Sci Rep 7:11553
Bourne DG, Morrow KM, Webster NS (2016) Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu Rev Microbiol 70:317–340
Browne NK, Smithers SG, Perry CT (2013) Spatial and temporal variations in turbidity on two inshore turbid reefs on the Great Barrier Reef, Australia. Coral Reefs 32:195–210
Bruce T, Meirelles PM, Garcia G, Paranhos R, Rezende CE, de Moura RL, Coni EO, Vasconcelos AT, Amado Filho G, Hatay M (2012) Abrolhos bank reef health evaluated by means of water quality, microbial diversity, benthic cover, and fish biomass data. PLoS ONE 7:e36687
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621
Damjanovic K, Blackall LL, Menéndez P, van Oppen MJ (2020) Bacterial and algal symbiont dynamics in early recruits exposed to two adult coral species. Coral Reefs 39:189–202
De’ath G, Fabricius K (2010) Water quality as a regional driver of coral biodiversity and macroalgae on the Great Barrier Reef. Ecol Appl 20:840–850
DeSalvo MK, Sunagawa S, Fisher PL, Voolstra CR, Iglesias-Prieto R, Medina M (2010) Coral host transcriptomic states are correlated with Symbiodinium genotypes. Mol Ecol 19:1174–1186
Díaz JM, Barrios LM, Cendales MH, Garzón-Ferreira J, Geister J, López-Victoria M, Ospina GH, Parra-Velandia F, Pinzón J, Vargas-Angel B (2000) Áreas coralinas de Colombia. INVEMAR Ser Publicaciones Espec 5:176
DiPerna S, Hoogenboom M, Noonan S, Fabricius K (2018) Effects of variability in daily light integrals on the photophysiology of the corals Pachyseris speciosa and Acropora millepora. PloS One 13(9):e0203882
Erftemeijer PL, Hagedorn M, Laterveer M, Craggs J, Guest JR (2012a) Effect of suspended sediment on fertilization success in the scleractinian coral Pectinia lactuca. J Mar Biol Assoc UK 92:741–745
Erftemeijer PL, Riegl B, Hoeksema BW, Todd PA (2012b) Environmental impacts of dredging and other sediment disturbances on corals: a review. Mar Pollut Bull 64:1737–1765
Fisher R, Bessell-Browne P, Jones R (2019) Synergistic and antagonistic impacts of suspended sediments and thermal stress on corals. Nat Commun 10:2346
Gardner TA, Côté IM, Gill JA, Grant A, Watkinson AR (2003) Long-term region-wide declines in Caribbean corals. Science 301:958–960
Ginestet C (2011) ggplot2: elegant graphics for data analysis. J R Stat Soc Ser A Stat Soc 174:245–246
Guest JR, Edmunds PJ, Gates RD, Kuffner IB, Andersson AJ, Barnes BB, Chollett I, Courtney TA, Elahi R, Gross K (2018) A framework for identifying and characterising coral reef “oases” against a backdrop of degradation. J Appl Ecol 55:2865–2875
Haas AF, Fairoz MF, Kelly LW, Nelson CE, Dinsdale EA, Edwards RA, Giles S, Hatay M, Hisakawa N, Knowles B (2016) Global microbialization of coral reefs. Nat Microbiol 1:16042
Hoegh-Guldberg O (2011) Coral reef ecosystems and anthropogenic climate change. Reg Environ Change 11:215–227
Hoegh-Guldberg O, Mumby PJ, Hooten AJ, Steneck RS, Greenfield P, Gomez E, Harvell CD, Sale PF, Edwards AJ, Caldeira K et al (2007) Coral reefs under rapid climate change and ocean acidification. Science 318:1737–1742
Hughes TP, Baird AH, Bellwood DR, Card M, Connolly SR, Folke C, Grosberg R, Hoegh-Guldberg O, Jackson JB, Kleypas J (2003) Climate change, human impacts, and the resilience of coral reefs. Science 301:929–933
Iglesias-Prieto R, Trench RK (1994) Acclimation and adaptation to irradiance in symbiotic dinoflagellates. I. Responses of the photosynthetic unit to changes in photon flux density. Mar Ecol Prog Ser Oldendorf 113:163–175
Iluz D, Alexandrovich I, Dubinsky Z (2012) The enhancement of photosynthesis by fluctuating light. Artif Photosynth 115–134
Jackson EJ, Donovan M, Cramer K, Lam V (2014) Status and trends of Caribbean coral reefs: 1970–2012. 306
Jeffrey SW, Humphrey GF (1975) New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem Physiol Pflanz 167:191–194
Jessen C, Lizcano JFV, Bayer T, Roder C, Aranda M, Wild C, Voolstra CR (2013) In-situ effects of eutrophication and overfishing on physiology and bacterial diversity of the Red Sea Coral Acropora hemprichii. PLoS ONE 8:e62091
Kaczmarsky LT, Draud M, Williams EH (2005) Is there a relationship between proximity to sewage effluent and the prevalence of coral disease. Caribb J Sci 41:124–137
Kemp DW, Hernandez-Pech X, Iglesias-Prieto R, Fitt WK, Schmidt GW (2014) Community dynamics and physiology of Symbiodinium spp. before, during, and after a coral bleaching event. Limnol Oceanogr 59:788–797
Klaus JS, Janse I, Heikoop JM, Sanford RA, Fouke BW (2007) Coral microbial communities, zooxanthellae and mucus along gradients of seawater depth and coastal pollution. Environ Microbiol 9:1291–1305
Kline DI, Kuntz NM, Breitbart M, Knowlton N, Rohwer F (2006) Role of elevated organic carbon levels and microbial activity in coral mortality. Mar Ecol Prog Ser 314:119–125
Kushmaro A, Banin E, Loya Y, Stackebrandt E, Rosenberg E (2001) Vibrio shiloi sp. nov., the causative agent of bleaching of the coral Oculina patagonica. Int J Syst Evol Microbiol 51:1383–1388
Lahti L, Shetty S et al (2017) Tools for microbiome analysis in R. Microbiome package version 1.7.2. Bioconductor
LaJeunesse T (2002) Diversity and community structure of symbiotic dinoflagellates from Caribbean coral reefs. Mar Biol 141:387–400
Lajeunesse TC, Trench RK (2000) Biogeography of two species of Symbiodinium (Freudenthal) inhabiting the intertidal sea anemone Anthopleura elegantissima (Brandt). Biol Bull 199:126–134
Lee ST, Davy SK, Tang S-L, Fan T-Y, Kench PS (2015) Successive shifts in the microbial community of the surface mucus layer and tissues of the coral Acropora muricata under thermal stress. FEMS Microbiol Ecol 91:fiv142
Lesser MP, Bythell JC, Gates RD, Johnstone RW, Hoegh-Guldberg O (2007) Are infectious diseases really killing corals? Alternative interpretations of the experimental and ecological data. J Exp Mar Biol Ecol 346:36–44
Lewis AM, Chan AN, LaJeunesse TC (2019) New species of closely related endosymbiotic dinoflagellates in the greater Caribbean have niches corresponding to host coral phylogeny. J Eukaryot Microbiol 66:469–482
Lima LF, Weissman M, Reed M, Papudeshi B, Alker AT, Morris MM, Edwards RA, de Putron SJ, Vaidya NK, Dinsdale EA (2020) Modeling of the coral microbiome: the influence of temperature and microbial network. Mbio 11
López-Victoria M, Rodríguez-Moreno M, Zapata FA (2015) A paradoxical reef from Varadero, Cartagena Bay, Colombia. Coral Reefs 34:231–231
Mahmoud HM, Kalendar AA (2016) Coral-associated Actinobacteria: diversity, abundance, and biotechnological potentials. Front Microbiol 7:204
McDaniel LD, Young E, Delaney J, Ruhnau F, Ritchie KB, Paul JH (2010) High frequency of horizontal gene transfer in the oceans. Science 330:50–50
McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217
Morgan KM, Perry CT, Johnson JA, Smithers SG (2017) Nearshore turbid-zone corals exhibit high bleaching tolerance on the Great Barrier Reef following the 2016 ocean warming event. Front Mar Sci 4:224
Morrow KM, Moss AG, Chadwick NE, Liles MR (2012) Bacterial associates of two Caribbean coral species reveal species-specific distribution and geographic variability. Appl Env Microbiol 78:6438–6449
Mumby PJ, Hastings A, Edwards HJ (2007) Thresholds and the resilience of Caribbean coral reefs. Nature 450:98
Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH (2007) The vegan Package. Community Ecol Package 10:631–637
Osinga R, Iglesias-Prieto R, Enriquez S (2012) Measuring photosynthesis in symbiotic invertebrates: a review of methodologies, rates and processes. In: Najafpour MM (ed) Applied photosynthesis. InTech, Rijeka, Croatia, pp 229–256. https://doi.org/10.5772/29339
Pandolfi JM, Bradbury RH, Sala E, Hughes TP, Bjorndal KA, Cooke RG, McArdle D, McClenachan L, Newman MJ, Paredes G (2003) Global trajectories of the long-term decline of coral reef ecosystems. Science 301:955–958
Peixoto RS, Rosado PM, de Leite DCA, Rosado AS, Bourne DG (2017) Beneficial microorganisms for corals (BMC): proposed mechanisms for coral health and resilience. Front Microbiol 8:341
Pinzón JH, Kamel B, Burge CA, Harvell CD, Medina M, Weil E, Mydlarz LD (2015) Whole transcriptome analysis reveals changes in expression of immune-related genes during and after bleaching in a reef-building coral. R Soc Open Sci 2:140214
Pizarro V, Rodríguez SC, López-Victoria M, Zapata FA, Zea S, Galindo-Martínez CT, Iglesias-Prieto R, Pollock J, Medina M (2017) Unraveling the structure and composition of Varadero Reef, an improbable and imperiled coral reef in the Colombian Caribbean. PeerJ 5:e4119
Pollock FJ, McMinds R, Smith S, Bourne DG, Willis BL, Medina M, Thurber RV, Zaneveld JR (2018) Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat Commun 9:1–13
Restrepo JC, Escobar J, Otero L, Franco D, Pierini J, Correa I (2016) Factors influencing the distribution and characteristics of surface sediment in the Bay of Cartagena, Colombia. J Coast Res 135–148
Restrepo JC, Escobar J, Otero L, Franco D, Pierini J, Correa I (2017) Factors influencing the distribution and characteristics of surface sediment in the Bay of Cartagena, Colombia. J Coast Res 33:135–148
Restrepo JD, Escobar R, Tosic M (2018) Fluvial fluxes from the Magdalena River into Cartagena Bay, Caribbean Colombia: trends, future scenarios, and connections with upstream human impacts. Geomorphology 302:92–105
Röthig T, Ochsenkühn MA, Roik A, Van Der Merwe R, Voolstra CR (2016) Long-term salinity tolerance is accompanied by major restructuring of the coral bacterial microbiome. Mol Ecol
Rutherford ST, Bassler BL (2012) Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med 2:a012427
Sarris PF, Trantas EA, Mpalantinaki E, Ververidis F, Goumas DE (2012) Pseudomonas viridiflava, a multi host plant pathogen with significant genetic variation at the molecular level. PLoS ONE 7
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60
Shaver EC, Shantz AA, McMinds R, Burkepile DE, Vega Thurber RL, Silliman BR (2017) Effects of predation and nutrient enrichment on the success and microbiome of a foundational coral. Ecology 98:830–839
Shlesinger T, Loya Y (2019) Breakdown in spawning synchrony: a silent threat to coral persistence. Science 365:1002–1007
Sunagawa S, DeSantis TZ, Piceno YM, Brodie EL, DeSalvo MK, Voolstra CR, Weil E, Andersen GL, Medina M (2009) Bacterial diversity and White Plague Disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J 3:512
Sutherland KP, Porter JW, Turner JW, Thomas BJ, Looney EE, Luna TP, Meyers MK, Futch JC, Lipp EK (2010) Human sewage identified as likely source of white pox disease of the threatened Caribbean elkhorn coral, Acropora palmata. Environ Microbiol 12:1122–1131
Sweet M, Bythell J (2015) White syndrome in Acropora muricata: nonspecific bacterial infection and ciliate histophagy. Mol Ecol 24:1150–1159
Sweet MJ, Brown BE (2016) Coral responses to anthropogenic stress in the twenty-first century: an ecophysiological perspective. In: Oceanography and marine biology. CRC Press, pp 279–322
Sweet M, Croquer A, Bythell J (2012) Dynamics of bacterial community development in the reef coral Acropora muricata following experimental antibiotic treatment. Coral Reefs 30
van Oppen MJ, Blackall LL (2019) Coral microbiome dynamics, functions and design in a changing world. Nat Rev Microbiol 17:557–567
Van Woesik R, Houk P, Isechal AL, Idechong JW, Victor S, Golbuu Y (2012) Climate-change refugia in the sheltered bays of Palau: analogs of future reefs. Ecol Evol 2:2474–2484
Vega L, Ordoñez JI, Pinilla G (2013) Towards a systemic assessment of environmental impact (SAEI) regarding alternative hydrosedimentological management practice in the Canal del Dique, Colombia. Ing E Investig 33:41–48
Vega Thurber RL, Burkepile DE, Fuchs C, Shantz AA, McMinds R, Zaneveld JR (2014) Chronic nutrient enrichment increases prevalence and severity of coral disease and bleaching. Glob Change Biol 20:544–554
Wagner DE, Kramer P, Van Woesik R (2010) Species composition, habitat, and water quality influence coral bleaching in southern Florida. Mar Ecol Prog Ser 408:65–78
Wang Z, Guo F, Liu L, Zhang T (2014) Evidence of carbon fixation pathway in a bacterium from candidate phylum SBR1093 revealed with genomic analysis. PLoS ONE 9
Wenger AS, Williamson DH, da Silva ET, Ceccarelli DM, Browne NK, Petus C, Devlin MJ (2016) Effects of reduced water quality on coral reefs in and out of no-take marine reserves. Conserv Biol 30:142–153
Williams AD, Brown BE, Putchim L, Sweet MJ (2015) Age-related shifts in bacterial diversity in a reef coral. PLoS ONE 10:e0144902
Wooldridge SA (2009) Water quality and coral bleaching thresholds: formalising the linkage for the inshore reefs of the Great Barrier Reef, Australia. Mar Pollut Bull 58:745–751
Zaneveld JR, Burkepile DE, Shantz AA, Pritchard CE, McMinds R, Payet JP, Welsh R, Correa AM, Lemoine NP, Rosales S (2016) Overfishing and nutrient pollution interact with temperature to disrupt coral reefs down to microbial scales. Nat Commun 7:11833
Zaneveld JR, McMinds R, Thurber RV (2017) Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat Microbiol 2:17121
Ziegler M, Seneca FO, Yum LK, Palumbi SR, Voolstra CR (2017) Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat Commun 8:14213
Ziegler M, Grupstra CG, Barreto MM, Eaton M, BaOmar J, Zubier K, Al-Sofyani A, Turki AJ, Ormond R, Voolstra CR (2019) Coral bacterial community structure responds to environmental change in a host-specific manner. Nat Commun 10:3092
Acknowledgements
This study was funded by the National Science Foundation (Grant NSF OCE 1642311 to M. M. and R. I. P, and NSF OCE 1442206 to M. M) and Penn State grants from the Institute for Energy and the Environment and the Social Sciences Institute to M.M. We want to thank professors Gabriel Navas, Adriana Bermúdez and Dario Mendez from Universidad de Cartagena for their hospitality and support at their campus. We also want to thank the personnel of the Corales del Rosario y San Bernardo National Natural Park, in particular to Esteban Zarza, for their constant support to our projects within the park. The light system for PE curves was designed and made by Miguel A. Gómez-Reali from Universidad Nacional Autónoma de México UNAM. Lastly, we would like to thank the dive shops Cartagena Divers and Scuba Cartagena, as well as Pablo and Hector Avendaño from Bocachica (Tierra Bomba island) for their support during field work. The research was conducted under the collection permit No. 0546 from 2014 issued by the “Autoridad Nacional de Licencias Ambientales ANLA”.
Author information
Authors and Affiliations
Contributions
MM and RIP conceived of and, along with FJP, designed the study. FJP, TLL, CTGM, KGC, LAGG, MLV, and VP ran/participated heavily in field work and data collection. SR participated in field work and data collection, performed statistical analyses, and wrote the manuscript. MM, TLL, and MLV supported writing and editing of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicting interests.
Additional information
Topic Editor Kyle Morgan
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
16S data has been submitted to ENA under Submission ID: SUB7604577 and BioProject ID: PRJNA639618
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure
1. Bar plot showing relative survivorship for all transplanted fragments 6 months after the transplant. All fragments moved to/within Varadero survived, and Cartagena Bay had the highest overall fragment mortality. No significant differences in mortality were found between fragments based on origin site or transplant site (p = 0.148; Kruskal–Wallis rank-sum test) (PNG 98 kb)
Figure
2. Principal coordinates analysis (PCoA) of Bray–Curtis matrices of the microbial communities associated with samples from different transplant time points and treatments alongside 7 water samples (n = 3 Rosario, n = 3 Varadero, and n = 1 Cartagena Bay). Water samples were compared to (a) all fragment samples across all time points, and (b) only post-transplant fragments (PNG 145 kb)
Figure
3. Family level bar plot of the top 12 most abundant OTUs in pre-transplant fragments, calculated by origin site. “Low abundance” category contains all OTUs not in the top 12 (PNG 90 kb)
Figure
4. Violin alpha diversity plot (Chao1) for all post-transplant fragments by transplant site. Cartagena Bay fragments had significantly higher diversity than Rosario or Varadero fragments (Wilcoxon rank-sum test, p < 0.0001) (PNG 43 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Roitman, S., López-Londoño, T., Joseph Pollock, F. et al. Surviving marginalized reefs: assessing the implications of the microbiome on coral physiology and survivorship. Coral Reefs 39, 795–807 (2020). https://doi.org/10.1007/s00338-020-01951-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00338-020-01951-5