Abstract
The AP2/ERF transcription factor family plays an important role in different biological processes such as growth, development and response to abiotic and biotic stresses in plants. The genome-wide analysis identified 531 AP2/ERF genes in Brassica napus (oilseed rape or canola) that ranged from 333 to 6440 bp in genomic and 273–2493 bp in coding DNA sequence length. We classified BnAP2/ERF proteins into five subfamilies including AP2 (58 genes), ERF (250 genes), DREB/CBF (194 genes), RAV (26 genes), and Soloist (3 genes). Furthermore, AP2/ERF proteins were subdivided into 15 groups according to the AP2/ERF classification in Arabidopsis. The number of exons in BnAP2/ERF genes was from one to eleven and most of these genes in the same subfamily had the same exon–intron pattern. The results also indicated that the composition of conserved motifs in most proteins in each group was similar. The intron–exon patterns and the composition of conserved motifs validated the BnAP2/ERF transcription factors phylogenetic classification. Based on the results of genome distribution, BnAP2/ERF genes were located unevenly on the 19 B. napus chromosomes. The results indicated that gene duplication may play an important role in genome expansion of B. napus. Furthermore, genome evolution of B. napus using orthologous and paralogous identification was studied. We found 278, 380 and 366 orthologous gene pairs between B. napus with A. thaliana, B. rapa and B. oleracea, respectively. The results of this study will be useful in investigation of functional role and molecular mechanisms of BnAP2/ERF transcription factors genes in response to different stresses.
Similar content being viewed by others
Introduction
Abiotic or biotic stresses such as drought, salinity, heat, cold, heavy metals, oil contamination, nematodes, fungus, and pathogens influence growth and productivity of crops (Bartels and Sunkar 2005; Rastgoo et al. 2011; Mojarad et al. 2016; Ghoreishi et al. 2017). Plants use different mechanisms at physiological, biochemical, and molecular levels to increase tolerance to these stresses (Suzuki et al. 2014; Sami and Alemzadeh 2016; Haghir and Alemzadeh 2018; Atashi Shirazi et al. 2019). Plants respond to different stresses by regulation of stress-responsive genes expression through transcription factors (TFs) (Yamaguchi-Shinozaki and Shinozaki 2006; Olfatmiri et al. 2014). Different transcription factor families have been identified in plants such as AP2/ERF, Myb, Hsp, bZIP, NAC, Dof, MADS-box, etc. (Yamaguchi-Shinozaki and Shinozaki 2006).
The APETALA 2/Ethylene-Responsive Factor (AP2/ERF) family is a large family of transcription factors in plants, which is characterized by the presence of at least one AP2 DNA-binding domain consisting of a highly conserved sequence about 60 amino acids (Riechmann and Meyerowitz 1998). ERF and AP2 subfamilies consist of one and two AP2 domains, respectively, although there are some proteins with one domain which are classified in AP2 group (Riechmann and Meyerowitz 1998; Licausi et al. 2013). AP2 subfamily, based on amino acid sequence of AP2 domain and nuclear localization sequence can be classified into AP2 and ANT groups (Shigyo and Ito 2004). RAV (Related to ABI3 and VP1) subfamily contains two different DNA-binding domains, one AP2 domain and one additional B3 domain. There is another subfamily with a single AP2 domain called soloist which differs from ERF subfamily due to its sequence and gene structure (Nakano et al. 2006). Sakuma et al. (2002) reported 145 genes as members of AP2/ERF family in Arabidopsis and based on the similarity of the amino acid sequences in the AP2 domains of AtAP2/ERF, classified them into five groups: AP2 subfamily (17 members, 14 genes with double AP2 domains and 3 genes with single AP2 domain), ERF subfamily (65 members), DREB/CBF (Dehydration-Responsive Element Binding-proteins/C-repeat Binding Factor) subfamily (56 members), RAV subfamily (6 members), and unclassified AL079349 gene, which further Nakano et al. (2006) explained this gene is identical to the soloist gene AT4g13040 in their study. Nakano et al. (2006) found 147 AP2/ERF transcription factor genes in Arabidopsis and grouped them into AP2 subfamily (18 genes, 14 with double AP2 domains and 4 with single AP2 domain), ERF subfamily (ERF and DREB/CBF together, 122 genes), RAV subfamily (6 genes), and AT4g13040 as soloist. ERF subfamily in their study was divided into 12 groups based on gene structures and conserved motifs.
One of the most important members of Brassicaceae family is B. napus (oilseed rape or canola), which is grown worldwide especially as oilseed crop, and is also an important crop for biodiesel, biofuel, and pharmaceutical products. B. napus (genome AACC, 2n = 4x = 38) is an allopolyploid which was produced by hybridization and chromosome doubling between Brassica rapa (Chinese cabbage, genome AA, 2n = 2x = 20) and Brassica oleracea (Mediterranean cabbage, genome CC, 2n = 2x = 18) about 7500 years ago (Chalhoub et al. 2014). Song et al. (2016), described the origination, functional characterization, and evolutionary pattern of the AP2/ERF superfamily and identified 515 AP2/ERF genes in B. napus. Owji et al. (2017) identified 321 AP2/ERF genes in B. napus. In this study, we described the AP2/ERF transcription factors in B. napus through a genome-wide analysis. We found 531 AP2/ERF genes in B. napus and grouped them according to the AP2/ERF classification in Arabidopsis (Nakano et al. 2006). Identification and characterization of AP2/ERF transcription factor family in B. napus could help us to elucidate molecular mechanisms involved in response to biotic and abiotic stresses.
Materials and methods
Identification of AP2/ERF family proteins in B. napus
To identify AP2/ERF family in canola, annotated proteins of B. napus were retrieved from the Genoscope database (http://www.genoscope.cns.fr). HMMER software version 3.0 (http://hmmer.janelia.org/) with the default parameters, was used for identification of AP2/ERF proteins using profile hidden Markov model. As a query AP2 domain (PF00847) was downloaded from the Pfam database (http://pfam.xfam.org). SMART (http://smart.embl-heidelberg.de/) and Pfam databases were used to confirm the presence of AP2 domain in each protein.
Multiple-sequence alignment, and phylogenetic analysis
Multiple sequence alignment of AP2 domain sequences was performed using ClustalW in MEGA6.0 software (http://www.megasoftware.net/). A phylogenetic tree was generated by the neighbor-joining (NJ) method with the following parameters: Poisson correction, pairwise deletion option and a bootstrap test with 1000 replications to define the reliability of the resulting tree. The numbers indicated for each clade represent percentage of bootstrap values.
Determination of conserved motifs and gene structure analysis
Conserved motifs were identified by online MEME (http://meme.sdsc.edu/meme/meme.html) program using the full-length protein sequences of each AP2/ERF family member, with the following parameters: maximum number of motifs (30 motifs) and motif width set as 6–100 amino acid. For gene structure analysis, coding sequences and genomic sequences of each AP2/ERF were obtained from Genoscope database (http://www.genoscope.cns.fr) and then exon–intron structures were drawn using Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/).
Chromosomal locations of BnAP2/ERF genes
The chromosomal positions of all AP2/ERF genes were retrieved from B. napus information resource database (http://www.genoscope.cns.fr.). The physical distribution of AP2/ERF genes was drawn with MapChart v.2.2 and the BnAP2/ERF genes were graphically displayed on the B. napus chromosomes.
Identification of orthologous and paralogous BnAP2/ERF genes between three species of brassica and Arabidopsis thaliana
The orthologous genes among B. napus, B. rapa, B. oleracea and Arabidopsis thaliana were identified from EnsemblPlants (http://plants.ensembl.org/index.html). When the identity exceeded 70%, it was considered to indicate orthologous genes. The paralogous genes in BnAP2/ERF proteins were identified with identity more than 85% from EnsemblPlants. Orthologous and paralogous BnAP2/ERF genes were displayed using Circos program (http://mkweb.bcgsc.ca/tableviewer/visualize/).
Gene ontology (GO) enrichment
GO enrichment analysis of the genes was conducted using the AgriGO platform, agriGO v2.0, (http://systemsbiology.cau.edu.cn/agriGOv2/) with default parameters like Fisher statistical test method, Multi-test adjustment method of Yekutieli (FDR under dependency) with significant level of 0.05 and complete GO gene ontology type (Tian et al. 2017). A Singular Enrichment Analysis (SEA) was performed using Arabidopsis genemodel (TAIR) as reference and FDR less than < 0.05.
Cis-element analysis
The 1500 bp upstream sequence of each gene, as promoter region, was extracted from ensemble plant (http://plants.ensembl.org). These sequences were analyzed using PlantCARE databases (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al. 2002).
Results and discussion
Identification of AP2/ERF transcription factor genes in B. napus
In total, 531 genes encoding AP2/ERF transcription factors were detected in B. napus genome. Song et al. (2016) previously identified 515 AP2/ERF genes in this species. These genes include complete or incomplete AP2 domain. After that, Owji et al. (2017) reported 321 genes as members of AP2/ERF transcription factors. In their study, only proteins contain complete AP2 domain assigned as AP2/ERF. In our study, 18 novel genes (BnaC07g50570D, BnaC09g24990D, BnaCnng49280D, BnaA08g30930D, BnaA08g00990D, BnaC03g69940D, BnaC05g51190D, BnaC01g03970D, BnaA07g12190D, BnaC07g48550D, BnaC06g03690D, BnaA06g02760D, BnaC06g03910D, BnaA06g02630D, BnaC06g03900D, BnaC06g04340D, BnaC05g27060D, and BnaA05g15500D) exceeded those of Song et al. (2016) and were detected as AP2/ERF superfamily; while, 2 genes of their study (BnaC09g28540D and BnaC03g77730D) did not identify in our study. A complete list of BnAP2/ERF genes and their detailed information such as gene stable ID, AP2/ERF family, chromosome number, gene start position, gene end position, gene length, CDS length, number of introns and number of exons are presented in the supplementary file: Table 1 S1 and S2.
Multiple sequence alignment and phylogenetic analysis of BnAP2/ERF transcription factors
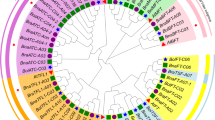
Multiple sequence alignment analysis was performed using conserved amino acid sequences of BnAP2 domains to detect the phylogenetic relationship between AP2/ERF transcription factors (supplementary file: Figure S1). Phylogenetic tree showed that BnAP2/ERF proteins, based on similarity in conserved amino acid sequences in AP2 domains and number of their DNA binding domains, may be classified in five subfamilies AP2, ERF, DREB/CBF, RAV, and Soloist (Fig. 1).
From 531 AP2/ERF transcription factors in B. napus, 58 proteins that were assigned to AP2 subfamily, 78% (45 proteins) had tandem repeated of AP2 domains, and just 22% (13 proteins) contained one AP2 domain. These 13 proteins were distinct from ERF subfamily and exhibited high similarity to other AP2 subfamily members, therefore these proteins were assigned to AP2 subfamily. Twenty-six proteins containing one AP2 domain together with B3 domain were classified as RAV subfamily. Four hundred forty-four proteins of BnAP2/ERF contained one AP2 domain and assigned to ERF subfamily. The members of ERF subfamily based on the similarity of AP2 domain sequence were classified into two groups of ERF subfamily (250 proteins) and DREB/CBF subfamily (194 proteins). These two subfamilies differ from each other at two conserved amino acid positions in AP2 domain, 14th and 19th residues. DREB subfamily proteins (group A) contained valine at position 14 (V14) and glutamic acid at position 19 (E19), whereas ERF subfamily proteins (group B) contained alanine at position 14 (A14) and aspartic acid at position 19 (D19); however, V14 is more important than E19 in detection of DREB subfamily (Sakuma et al. 2002; Nakano et al. 2006). The remaining three proteins, BnaC08g08940D, BnaA08g04940D, and BnaC08g08930D, contained one AP2 domain that showed low homology with other members of AP2/ERF transcription factors and high homology with Arabidopsis Soloist (At4g13040), therefore it was assigned as Soloist. The number of members in this subfamily was equal to what has been reported by Song et al. (2016) (supplementary file: Figure S1).
In the AP2/ERF family, YRG, WLG, AA, and RAYD elements were identified in AP2 domains. Based on sequence alignment of AP2 domains, WLG, AA, and AYD elements were highly conserved in all BnAP2/ERF proteins (supplementary file: Figure S2). In the RAV subfamily, WLG and AA elements were conserved in all sequences; however, YD residues of RAYD was highly conserved in this subfamily except two proteins which were converted to FD (supplementary file: Figure S2). In the soloist subfamily, WLG element was converted to HLG, AA element conserved in all sequences and RAYD element was converted to RLYD (supplementary file: Figure S2). AP2 subfamily contained two AP2 domains (AP2-R1 and AP2-R2). In this subfamily, AA element was conserved in most proteins in both domains; WLG element in AP2-R2 domain was converted to YLG in most proteins. RAYD element was conserved in AP2-R1 domain with the maximum number of members and in some other proteins was converted to RSYD or HTYD. However, in AP2-R2 domain, fewer members contained RAYD element and in most proteins, this element was converted to EAYD whereas, in some members, it was converted to AAYD and IAYD (supplementary file: Figures S2 and S2). In DRREB/CBF subfamily, although, YRG element was conserved in most members but in some of them converted to YKG, FRG, HRG, YGG, or YHG. WLG and AA elements were highly conserved in all DREB/CBF subfamily members. RAYD element showed the highest variation in this subfamily; the majority of proteins contained RAYD, LAYD, RAHD, or MAYD sequences, and some of them contained SAYD, AAYD, VAHD, or RAFD sequences. Also, the results showed that LNFP element was highly conserved and detected in the C-terminal region of AP2 domain in most members of this subfamily (supplementary file: Figure S2). All members of this subfamily had valine residue in the position 14th (V14), except BnaC07g50570D, BnaCnng49280D, and BnaA08g30930D proteins which had incomplete AP2 domain. The majority of proteins in this subfamily had glutamic residue at position 19th (E19). Another conserved element in this subfamily was AEIR element, although in some members, it converted to SEIR and CEVR sequences. All proteins in DREB/CBF subfamily contained alanine at position 37 (A37).
It has been previously demonstrated that the formation of an α-helix in this position is essential for binding to DRE element and GCC box in the promoter of many abiotic and biotic stress-inducible genes (Liu et al. 2006; Allen et al. 1998). In the ERF subfamily, most members contained YRG element, but in some sequences, it was converted to FRG, HKG or FLG sequences. WLG, AA, and RAYD elements were conserved in majority of ERF subfamily proteins, but in some members, RAYD element was conserved to KAYD, LAYD, or MAYD sequences. Alanine at position 14 (A14) and aspartic acid at position 19 (D19) were conserved in most ERF subfamily members. Also, alanine residue at position 37 (A37) was detected in most proteins of this subfamily. In this subfamily, AEIR element was conserved in majority of proteins, while in some of them, it was converted to SEIR, AEIK or AEIW sequences. LNFP element was also conserved in the AP2 domain of most ERF subfamily proteins. In addition, some other elements such as TNFP and VNFP were also detected in these proteins (supplementary file: Figure S2).
Gene ontology
Gene ontology (GO) enrichment analysis was carried out to determine the role of AP2/ERF transcription factors in the biology of Brassica at the molecular, cellular and organism levels. Gene ontology analysis was carried out for 266 out of 531 AP2/ERF genes. Based on GO analysis, the genes encoding AP2/ERF transcription factors were categorized into 3 groups of biological process (BP), molecular function (MF), and cellular component (CC). The aforementioned groups, BP, MF and CC consisted of 18 (5:2:11:11:6:10:11:3:2:3:14:12:6:5:2:1:0:1 subcategories), 3 (6:5:1 subcategories) and 4 (4:1:2:1 subcategories) categories, respectively (Fig. 2 and supplementary file: Table 1 S3). As it was expected, all products of the genes were located within living cells, but interestingly, only the product of one of them, BnaC03g69940D, was associated with plasma membrane. This protein belongs to group VII of ERF subfamily which has CMVII-1 motif with the sequence of MCGGAII in the N-terminal region. This protein has also the CMVII-4 motif with the sequence of TPEISS which is phosphorylated by MAPK. The results also showed that all of these proteins associated with organelles such as nucleus.
The molecular functions of these proteins have been also determined. The results of GO analysis showed that all these proteins were located in the transcription regulator activity category (GO:0140110) and the subcategory of DNA-binding transcription factor activity (GO:0003700). It was expected since these proteins are transcription factors, and this result confirmed the accuracy of the collected sequences. Seven of these proteins, BnaA06g33420D (belong to RAV subfamily), BnaC04g25670D (belong to group VII of ERF subfamily), BnaA02g27630D (belong to RAV subfamily), BnaA09g30810D (belong to group I of DREB subfamily), BnaA01g04150D (belong to group III of DREB subfamily), BnaA10g25000D (belong to group IV of DREB subfamily), and BnaAnng13220D (belong to group IV of DREB subfamily) located in the category of binding (GO:0005488) that indicate these proteins have the interaction with one or more specific sites on another molecule. Among these proteins, 6 proteins interact with an organic heterocyclic compound and only one protein, BnaA06g33420D, related to RAV subfamily does not interact with organic heterocyclic compounds. BnaA06g33420D and BnaC04g25670D are the only proteins in this category that interact with ions. But, the most interesting protein in this category is BnaC04g25670D, a protein belonging to group IX of ERF subfamily that located in all subcategories of GO:0005488. This protein interacts with organic heterocyclic compounds, carbohydrate derivatives, some small molecules, ions, and even drugs. It indicated that this protein may be a critical molecule. So, the gene encoding this protein was selected for more cis element analysis. Three proteins BnaA01g06090D (belong to group II of DREB subfamily), BnaC04g25670D (belong to group IX of ERF subfamily), and BnaA06g33420D (belong to RAV subfamily) have catalytic activity and five enzymatic activities have been figured out for them. BnaA01g06090D has a transferase activity and includes a BPL/LPL catalytic domain which catalyze the covalent attachment of biotin and lipoic acid to various multicomponent enzyme complexes that catalyze key metabolic reactions (Reche 2000). It was cleared BnaC04g25670D that interacts with various molecules also have ligase activity and catalytic activity that acts to modify RNA. Bra025159 is the third protein in this group that has catalytic activity and catalyze the hydrolysis of proteins. These protein in addition to AP2 and B3 domain has a peptidase M24 (methionine aminopeptidase, suerfamily1) domain that catalyses the hydrolytic cleavage of the N-terminal methionine from newly synthesised polypeptides (Xiao et al. 2010). In addition to Bra027447, the genes encoding Bra011244 and Bra025159 were also selected for more cis element analysis.
Another group of GO analysis is BP in which the contribution of each protein in different processes was determined. The result showed that the products of four genes including BnaA03g24540D, BnaA02g08130D, BnaA02g14040D, and BnaC04g25670D are involved in localization (GO:0051179). One of these proteins, BnaA03g24540D, involved in all subcategories in this field. BnaC04g25670D also plays a role in the establishment of localization to localize a substance or cellular component. Response to biotic and abiotic stimulus is an important biological process and the most important 11 proteins in this category were BnaA08g04090D (response to cold stress); BnaA03g24540D and BnaA02g08130D (response to freezing stress); BnaA10g25000D, and BnaA01g09230D (response to heat stress); BnaAnng13220D, BnaA06g10760D, BnaA06g02670D, and BnaAnng03730D (response to salinity and osmatic stress); BnaA01g09230D (response to reactive oxygen species, ROS stress); BnaA10g25000D, BnaA08g04090D, BnaA06g02670D, BnaA06g10760D, and BnaAnng03730D (response to water deficiency stress); BnaAnng21280D, and BnaAnng03730D (response to abiotic stress), and BnaA10g05780D (mechanical stress like wounding). BnaAnng21280D is also involved in jasmonic acid pathway, in addition to response to abiotic stress. In addition, we found that most proteins including BnaA03g24540D, BnaA02g08130D, BnaA10g25000D, and BnaA01g09230D that were involved in cold and freezing stress, also involved in response to light stimulus. Also, we found most proteins, including BnaA10g25000D, BnaA08g04090D, BnaA06g02670D, and BnaAnng03730D, involved in response to water deficiency stress, also involved in some other abiotic stresses such as cold, heat, salinity, and osmatic stress. GO analysis also showed that among these 11 proteins, only BnaAnng03730D was involved in biotic and abiotic stress. These proteins were also selected for cis element analysis.
Cis-element analysis
The selected genes based on GO analysis, were analyzed to find out cis elements at promoter region. The selected genes belong to various subfamilies, DREB (5 members,), ERF (5 members), AP2 (2 members), and RAV (1 member). The results showed the presence of ABRE element, in the promoter region of all selected genes. This element is a cis-acting element involved in abscisic acid responsiveness (Tuteja 2007). It has been previously reported that some transcription factors regulate ABA-dependent gene expression through interacting with ABRE element (Tuteja 2007). The copy number of this element vary from 1 to 4 and the position of this element was different from 53 to 1365 in their promoter regions. The member of RAV subfamily, BnaA06g33420D, has only one copy of this element at position 1365. Also, 3 members from ERF subfamily, BnaA01g09230D, BnaA10g05780D, and BnaA03g24540D, and one member from DREB subfamily, BnaA08g04090D, have only one copy of this element in their promoter regions. In addition, we found DRE element in the promoter regions of two members of ERF subfamily, BnaA06g02670D and BnaA03g24540D, that has been previously shown to be involved in abiotic stress (Tuteja 2007). BnaA03g24540D has only one copy of DRE element in its promoter region, and the obtained results from GO analysis indicated that its product has a role in light and freezing stress. It has been previously reported that DRE element is involved in light and cold pathways (Franklin 2009). BnaA06g02670D has two copies of DRE element in its promoter region and the results of GO analysis figured out that its product play a role in water deficiency, salinity and osmotic stress. The sequences of this copies are not exactly the same, one of them at position 176 (ACCGAGA) has been reported to mediate an ABA-dependent gene regulation in response to water deficiency stress, and the other one at position 431 (GCCGAC) shown involved in ABA-dependent response to osmotic stress (Roychoudhury et al. 2013). It showed that cis element analysis results corresponds to the results of GO analysis that was confirmed by previous studies.
Identification and characterization functional elements in DREB/CBF and ERF subfamilies
Functional elements outside DNA-binding domain regions in transcription factors play important roles in the function of these proteins such as transcriptional activity, protein–protein interactions, and nuclear localization (Liu et al. 1999). Identification of elements in each group can help to identify probable functional role of unknown TFs proteins, due to the fact that proteins in each group have almost similar functions (Nakano et al. 2006). A multiple sequence alignment using complete amino acid sequences of each group was performed to investigate functional elements features in DREB/CBF and ERF subfamilies. Therefore, ERF and DREB subfamilies were subdivided into 12 groups of I to X-L according to the grouping in Arabidopsis and rice (Nakano et al. 2006; Sakuma et al. 2002). DREB subfamily was clustered into 4 groups (I, II, III, IV) and ERF subfamily clustered into 8 groups [V, VI, VI-Like (VI-L), VII, VIII, IX, X and X-Like (X-L)] (supplementary file: Figure S3 and Table 1 S4).
Based on the results of multiple sequence alignment, two motifs, CMI-1 and CMI-2, were detected in some members of group I in DREB subfamily. It has been previously demonstrated that maize DBF1 contains these motifs; an additional effect on promoter activity was observed in the treatment of transgenic calli of maize were co-transferred with constructs containing Rab17 promoter and DBF1 with ABA (Kizis and Pages 2002). All members of group II in DREB subfamily contain CMII-1 motif in their C-terminal region adjacent to the AP2 domain. In addition, LWSY motif (CMII-3) was detected in the C-terminal region of some members of this group. Some members of this group also contained EAR motif (CMII-2) with consensus sequence of DLNxxP in the C-terminal region. EAR motif (ERF-associated Amphiphilic Repression) acts as a repressor domain in the repressor-type of AP2/ERF proteins (Fujimoto et al. 2000; Ohta et al. 2001). It has been also reported that AP2/ERF proteins containing EAR element, are involved in response to various biotic and abiotic stresses such as drought, cold, UV, hormone signaling and pathogen infection (Song et al. 2005; McGrath et al. 2005). CMIII-1 was identified in all proteins of group III in DREB subfamily in the C-terminal region after AP2 domain whereas, some of the proteins in this group had LWSY motif (CMIII-4) at the end of C-terminal region. Also, CMIII-3 motif with two ETRHP and DSAWR regions in both sides of AP2 domain was recognized in some of the proteins in this group that has been previously reported in the proteins of subgroup IIIc in Arabidopsis (Nakano et al. 2006). It has been shown that the members of subgroup IIIc were involved in the expression of genes in response to low temperature, drought, and salinity stresses (Liu et al. 1998; Gilmour et al. 2000; Haake et al. 2002). All proteins in group IV in DREB subfamily contained CMIV-1 in the N-terminal region before AP2 domain. AtDREB2A and AtDREB2B proteins are the members of group IV in Arabidopsis which have CMIV-1, CMIV-2 and CMIV-3 motifs (Nakano et al. 2006). It has been shown that these proteins have roles in DRE-dependent transcription (Liu et al. 1998).
Some proteins of group V in ERF subfamily had CMV-1 and CMV-2 motifs. It has been previously reported that some proteins like WIN1/SHN1 containing these motifs play an important role in increasing the accumulation of epidermal wax (Aharoni et al. 2004; Broun et al. 2004). The members of group VI ERF subfamily contained CMVI-1/CRF (cytokinin response factors) domain with ATDSS consensus sequence in the N-terminal region. Previous researches investigated that this domain has important roles in abiotic stresses such as cold and salinity stresses (Brenner et al. 2012). Some members of this group also contained CMVI-3 motif with consensus sequence of SP(T/V)SVL which is phosphorylated by mitogen-activated protein (MAP) kinase and/or casein kinase I. Tsi1 and Pti6 proteins from tobacco and tomato, respectively, contain the motifs of group VI and are involved in the response mechanisms to biotic and abiotic stresses (Zhou et al. 1997; Park et al. 2001). The members of group VI-Like in ERF subfamily had incomplete AP2 domain due to having CMVI-1 and CMVI-2 motifs assigned as group VI-Like. Furthermore, the proteins of this group had CMVI-5 motif with LPDxDFxD consensus sequence. All proteins in group VII from ERF subfamily had CMVII-1 motif with consensus sequence of MCGGAxI in the N-terminal region. In addition, in the C-terminal region of some proteins of this group LWSY motif was identified. In some other members of this group, the CMVII-4 motif with consensus sequence of TPxxxSS which is phosphorylated by MAPK was identified. Some proteins of group VIII in ERF subfamily had CMVIII-1 motif in the C-terminal region. It has been previously shown that proteins with CMVIII-1 and CMVIII-2 motifs in tobacco, Arabidopsis and rice play important roles in repressing of GCC box-mediated transcription (Fujimoto et al. 2000; Ohta et al. 2000, 2001). There were some proteins in group IX from ERF subfamily with the CMIX-5 motif in the C-terminal region which is phosphorylated by MAPK. It has been suggested that the members of this group are involved in response to different biotic stresses and phytohormones such as ethylene, jasmonate and salicylic acid (Gu et al. 2000, 2002; Berrocal-Lobo et al. 2002; Onate-Sanchez and Singh 2002). Proteins from group X in ERF subfamily had CMX-1 and CMX-2 motifs in the N-terminal region. The consensus sequence for CMX-2 motif is Cx2Cx4Cx2-4 that is located in the N-terminal region of proteins in groups X and X-Like. It was suggested that the repeated cysteine probably functions as a zinc-finger motif involved in DNA-binding or protein–protein interactions (Nakano et al. 2006). AtABR1, one of the members of group X in Arabidopsis, is involved in repressing ABA signaling pathway (Pandey et al. 2005). Some proteins in group X had an incomplete AP2 domain, therefore assigned as group X-Like. In addition to CMX-1 and CMX-2 motifs, CMX-3 motif was also identified in this group.
Analysis of gene structure and determination of conserved motifs in BnAP2/ERF transcription factors members
To study the gene structure, 531 BnAP2/ERF genes in the range 333 to 6440 bp in genomic and 273–2493 bp in coding DNA sequence (CDS) length were analyzed by Gene Structure Display Server (supplementary file: Figure S4). The number of exons in BnAP2/ERF genes was from one to eleven. The majority of genes (315 genes) were intronless and had no intron which includes about 60 percent of the total genes. These genes belonged to DREB, ERF and RAV subfamilies. DREB subfamily contained one to seven exons, while most genes in this subfamily (156 genes, about 80 percent) contained one exon. Only BnaA08g12710D, BnaA01g06090D, BnaC01g07340D and BnaC09g24990D genes, belonging to group II of DREB subfamily, contained 4 to 7 exons. The members of ERF subfamily contained 1 to 10 exons. BnaC06g33570D, BnaC04g25670D, BnaA05g25200D, BnaC05g31720D, BnaA05g18740D and BnaC01g40920D genes from this subfamily had 4 to 10 exons which belonged to groups VII, VIII and IX. Most number of exons, from 5 to 11, were observed in AP2 subfamily. BnaA05g02350D, BnaA10g21750D and BnaC04g02040D had 11 exons in this subfamily. RAV subfamily contained 1 to 3 exons except of BnaA06g33420D gene, which had 11 exons and 10 introns. In Soloist subfamily, BnaC08g08930D had 3 exons and BnaA08g04940D and BnaC08g08940D genes had 6 exons and 5 introns. The results of gene structure analysis were in agreement with the results of Song et al. (2016). Based on the result of gene structure analysis, most genes in the same subfamily had the same exon–intron pattern. Therefore, the results of gene structure analysis validated the results of phylogenetic classification.
The 30 conserved protein motifs were identified using MEME analysis of full-length protein sequences from BnAP2/ERF transcription factors members (supplementary file: Figure S4 and Table 1 S5). The conserved AP2 domain was predicted in motifs 1, 2, 3 and 4. These motifs were identified in most BnAP2/ERF transcription factors sequences. Motifs 5, 7, 16, 18, 20, 22 and 26 were identified in some proteins of AP2 family. Motif 6 was found in majority members of groups II and III in DREB subfamily. Motifs 8, 11, 14 and 21 were identified in some proteins of RAV family. Majority members of ERF and DREB subfamilies had motif 9 in their proteins. Also, this motif was found in two proteins of AP2 family, BnaA02g06490D and BnaA10g12950D which had one AP2 domain. Motif 10 was identified in 6 proteins of group VIII. Twenty-tree proteins of group III had motif 12 in their sequences. Motif 13 was identified in 11 members of group V. Majority of the genes of group VI had motif 15 in their sequences. Motif 17 was identified in 24 members of group IV. Some members of groups X and X-L had motifs 19 and 23 in their sequences, also motif 19 was found in BnaC06g10050D of group VIII. Motif 24 was identified in the members of groups VI and VI-L. Some members of groups VII and VIII had motif 25 in their sequences. Motif 27 was found in some proteins from different groups such as I, II, AP2, X, V, VI, IV and VIII. Five proteins of group X had motif 28 in their sequences. Motif 29 was identified in 5 proteins of group VIII. Motif 30 was found in some members of groups V, VI and IX. The composition of conserved motifs in most proteins in each group was similar, which validated the BnAP2/ERF transcription factors phylogenetic classification.
Chromosomal distribution of BnAP2/ERF genes
To determine the chromosomal distribution of BnAP2/ERF genes, each BnAP2/ERF gene was mapped on their corresponding chromosome according to gene information of B. napus database. The results showed that the BnAP2/ERF genes were distributed on all 19 chromosomes including 10 chromosomes related to sub-genome A and 9 chromosomes related to sub-genome C (Fig. 3). Two hundred sixty-two genes were located on sub-genome A chromosomes and 268 genes were located on sub-genome C chromosomes and only one gene (BnaUnng01150D), did not assign to any sub-genomes. The results indicated that genes 20, 28, 33, 11, 17, 19, 30, 27, 29, and 19 were located on chromosomes A1 to A10, respectively, and 29 BnAP2/ERF genes mapped onto unanchored chromosomes of sub-genome A (Ann chromosome). Also, genes 16, 25, 32, 24, 25, 28, 30, 25, and 26 were located on chromosomes C01 to C09, respectively and 37 genes mapped onto unanchored chromosomes of sub-genome C (Cnn chromosome). Chromosome A03 contained most AP2/ERF genes, and chromosome A04 contained the least of them. The members of AP2/ERF transcription factors families were unevenly located on each chromosome. AP2 subfamily members were distributed on all chromosomes except A04. Also, DREB and ERF subfamily members were located on all chromosomes. RAV subfamily members were found on A02, A05, A06, A08, A09, C02, C05, C06, and C07 chromosomes. Soloist subfamily members were located on A08 and C08 chromosomes.
Orthologous and paralogous genes study in BnAP2/ERF transcription factor
In this study, a comparative analysis was performed to identify orthologs of AP2/ERF genes between B. napus with Arabidopsis thaliana, B. rapa and B. oleracea genomes (Fig. 4, supplementary file: Table 1 S6, S7, S8). Based on the results, 265 genes in B. napus showed high similarity (identity > 70%) with 97 Arabidopsis genes and in total resulted in 278 orthologous gene pairs. In this study, each AtAP2/ERF gene had one to 6 orthologous genes in B. napus genome. It was previously suggested that each gene from A. thaliana had about six orthologous genes in B. napus (Wei 2007). Also, 380 orthologous gene pairs were identified between B. napus and B. rapa, according to 360 and 266 unique genes, respectively. Furthermore, 365 genes in B. napus showed high similarity with 264 B. oleracea genes resulting in 366 orthologous gene pairs. The number of orthologous gene pairs between B. napus with B. rapa and B. oleracea were higher than those of B. napus and A. thaliana. These results were almost consistent with the results of Song et al. (2016). They found 236, 425 and 429 orthologous gene pairs between B. napus with A. thaliana, B. rapa and B. oleracea, respectively. Furthermore, orthologous gene pairs 54 and 84 showed 100% identity between BnAP2/ERF genes with B. rapa and B. oleracea genes, respectively, while neither gene from A. thaliana showed 100% identity with BnAP2/ERF genes. B. napus (AACC) is a hybrid between B. rapa (AA) and B. oleracea (CC). Therefore, a stronger relationship was expected between B. napus with B. rapa and B. oleracea than A. thaliana. Orthologous genes between B. napus with B. rapa and B. oleracea suggesting whole genome duplication (polyploidy) plays an important role in the expansion of BnAP2/ERF genes.
Furthermore, 286 paralogous gene pairs (included 422 unique genes) with similarity greater than 85% were identified in BnAP2/ERF gene family (supplementary file: Table 1 S9, Fig. 5). These results indicated that gene duplication may play an important role in genome expansion. In the pathway, gene duplication consisted of tandem and chromosomal/segmental duplications. Distribution of BnAP2/ERF genes on 19 chromosomes showed that about 1.5% (8 of 531 total genes, 4 paired genes) of BnAP2/ERF genes were involved in tandem duplication with similarity greater than 90 percent. The pairs of tandem duplicated genes were BnaA04g14830D and BnaA04g14840D related to group X-L of ERF subfamily, BnaA08g30910D and BnaA08g30950D related to group III of DREB subfamily, on the A04 and A08 chromosomes respectively, and BnaC04g37670D and BnaC04g37680D related to group X-L of ERF subfamily, and BnaC08g08930D and BnaC08g08940D related to Soloist subfamily genes on the C04 and C08 chromosomes, respectively. In addition to tandem duplication in B. napus genome, chromosomal/segmental duplication was also identified. In this study, 282 paired genes (420 genes) were included in chromosomal/segmental duplication. Also, 96 paired genes showed high similarity (identity > 95%) in BnAP2/ERF genes. Most of them (91 paired genes) were between genome A and C, while 5 paired genes were within genome C chromosomes.
Based on the results of phylogenetic analysis, we classified BnAP2/ERF family proteins in five subfamilies, AP2 (58 members), ERF (250 members), DREB/CBF (194 members), RAV (26 members) and Soloist (3 members). Proteins of DREB/CBF and ERF were classified into four (I, II, III, IV) and eight (V, VI, VI-Like, VII, VIII, IX, X and X-Like) groups. BnAP2/ERF family proteins were about 3.6-fold more than Arabidopsis AP2/ERF (147 genes) (Nakano et al. 2006) and 1.8-fold more than B. rapa (291 genes) (Song et al. 2013). To date, number of reported AP2/ERF genes in B. napus were higher than of other plants (Song et al. 2016; Owji et al. 2017).
Based on the results of structure analysis and motif identification, different members of the same subfamily and group had similar gene structure and conserved protein motifs and therefore suggesting them to have same evolutionary origin and probably the same function. Orthologous genes are homologous genes descending from a common ancestor separated by a speciation event, while paralogous genes are homologue genes generating by a duplication event. Often orthologous genes have a similar function among different species, while paralogous have the same basic function, although they differ slightly in function. Therefore, the study of evolutionary genomics can shed light on the gene function. Due to close relationship between B. napus with A. thaliana, B. rapa and B. oleracea; homologous genes (identity > 70) were studied between them and 278, 380 and 366 orthologous gene pairs were identified, respectively. Also, 0, 54 and 84 highly similarity genes (identity 100%) were identified between B. napus with A. thaliana, B. rapa and B. oleracea, respectively. Due to the fact that B. rapa and B. oleracea are last ancestors of B. napus therefore, the number of orthologous genes and highly similarity genes between B. napus with B. rapa and B. oleracea was more than between B. napus and A. thaliana. Furthermore, paralogous genes in B. napus were studied and 286 paralogous gene pairs included 8 tandem duplicated genes and 282 chromosomal/segmental duplication paired genes were identified. Genome wide analysis of BnAP2/ERF genes showed that whole genome duplication, tandem duplication and chromosomal/segmental duplications play an important role in B. napus genome expansion. However, the number of tandem duplication was much lower than the number of whole genome duplication and chromosomal/segmental duplications, which showed these factors are main factors in evolution of BnAP2/ERF genes.
Conclusion
In this study, we identified 531 AP2/ERF transcription factors family genes in the Brassica napus genome. Different analysis such as, phylogenetic analysis and classification, conserved motif identification and gene structure analysis were performed on these genes. In addition, genome evolutionary was also studied. Analysis of AP2/ERF transcription factors genes can help to clarify biological functions and molecular mechanisms involved in response to different stresses. In addition, this comprehensive analysis provides a new achievement in development of beneficial traits in B. napus and other plants.
References
Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16:2463–2480
Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M (1998) A novel mode of DNA recognition by a b-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J 17:5484–5496
Atashi Shirazi F, Razi H, Niazi A, Alemzadeh A (2019) Molecular cloning and expression analysis of a stress-responsive WRKY transcription factor gene, BnWRKY57, from Brassica napus. Plant Omics J 12:37–47
Bartels D, Sunkar R (2005) Drought and salt tolerance in plants. Crit Rev Plant Sci 24:23–58
Berrocal-Lobo M, Molina A, Solano R (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29:23–32
Brenner WG, Ramireddy E, Heyl A, Schmulling T (2012) Gene regulation by cytokinin in Arabidopsis. Front Plant Sci 3:8. https://doi.org/10.3389/fpls.2012.00008
Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL (2004) WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Natl Acad Sci USA 101:4706–4711
Chalhoub B, Denoeud F, Liu S, Parkin IAP, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, Corréa M, Silva CD, Just J, Falentin C, Koh CS, Clainche IL, Bernard M, Bento P, Noel B, Labadie K, Alberti A, Charles M, Armaud D, Guo H, Daviaud C, Alamery S, Jabbari K, Zhao M, Edger PP, Chelaifa H, Tack D, Lassalle G, Mestiri I, Schnel N, Le Paslier MC, Fan G, Renault V, Bayer PE, Golicz AA, Manoli S, Lee TH, Thi VHD, Chalabi S, Hu Q, Fan C, Tollenaere R, Lu Y, Battail C, Shen J, Sidebottom CHD, Wang X, Canaguier A, Chauveau A, Bérard A, Deniot G, Guan M, Liu Z, Sun F, Lim YP, Lyons E, Town CD, Bancroft I, Wang X, Meng J, Ma J, Pires JC, King GJ, Brunel D, Delourme R, Renard M, Aury JM, Adams KL, Batley J, Snowdon RJ, Tost J, Edwards D, Zhou Y, Hua W, Sharpe AG, Paterson AH, Guan C, Wincker P (2014) Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345:950–953
Franklin KA (2009) Light and temperature signal crosstalk in plant development. Curr Opin Plant Biol 12:63–68
Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12:393–404
Ghoreishi G, Alemzadeh A, Mojarad M, Djavaheri M (2017) Bioremediation capability and characterization of bacteria isolated from petroleum contaminated soils in Iran. Sustain Environ Res 27:195–202
Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF (2000) Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 124:1854–1865
Gu YQ, Yang C, Thara VK, Zhou J, Martin GB (2000) Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 12:771–786
Gu YQ, Wildermuth MC, Chakravarthy S, Loh YT, Yang C, He X, Han Y, Martin GB (2002) Tomato transcription factors pti4, pti5, and pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 14:817–831
Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130:639–648
Haghir S, Alemzadeh A (2018) Cloning and molecular characterization of TaERF6, a gene encoding a bread wheat ethylene response factor. Mol Biol Res Commun 7:153–163
Kizis D, Pages M (2002) Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought-responsive element in an ABA-dependent pathway. Plant J 30:679–689
Lescot V, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Licausi F, Ohme-Takagi M, Perata P (2013) APETALA2/ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol 199:639–649
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391–1406
Liu L, White MJ, MacRae TH (1999) Transcription factors and their genes in higher plants functional domains, evolution and regulation. Eur J Biochem 262:247–257
Liu Y, Zhao TJ, Liu JM, Liu WQ, Yan YB, Zhou HM (2006) The conserved Ala37 in the ERF/AP2 domain is essential for binding with the DRE element and the GCC box. FEBS Lett 580:1303–1308
McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139:949–959
Mojarad M, Alemzadeh Am Ghoreishi G, Javaheri M (2016) Kerosene biodegradation ability and characterization of bacteria isolated from oil-polluted soil and water. J Environ Chem Eng 4:4323–4329
Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140:411–432
Ohta M, Ohme-Takagi M, Shinshi H (2000) Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J 22:29–38
Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13:1959–1968
Olfatmiri H, Alemzadeh A, Zakipour Z (2014) Up-regulation of plasma membrane H+-ATPase under salt stress may enable Aeluropus littoralis to cope with stress. Mol Biol Res Commun 4:65–73
Onate-Sanchez L, Singh KB (2002) Identification of Arabidopsis ethylene responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiol 128:1313–1322
Owji H, Hajiebrahimi A, Seradj H, Hemmati S (2017) Identification and functional prediction of stress responsive AP2/ERF transcription factors in Brassica napus by genome-wide analysis. Comput Biol Chem 71:32–56
Pandey GK, Grant JJ, Cheong YH, Kim BG, Li L, Luan S (2005) ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol 139:1185–1193
Park JM, Park CJ, Lee SB, Ham BK, Shin R, Paek KH (2001) Over-expression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13:1035–1046
Rastgoo L, Alemzadeh A, Afsharfifar A (2011) Isolation of two novel isoforms encoding zinc- and copper-transporting P1B -ATPase from Gouan (Aeluropus littoralis). Plant Omics J 4:377–383
Reche PA (2000) Lipoylating and biotinylating enzymes contain a homologous catalytic module. Protein Sci 9:1922–1929
Riechmann JL, Meyerowitz EM (1998) The AP2/EREBP family of plant transcription factors. Biol Chem 379:633–646
Roychoudhury A, Paul S, Basu S (2013) Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep 32:985–1006
Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290:998–1009
Sami Z, Alemzadeh A (2016) Isolation and molecular characterization of a novel Na+/H+ antiporter gene, AlNHX2, from Aeluropus littoralis and comparison of AlNHX1 and AlNHX2. Plant Omics J 9:205–212
Shigyo M, Ito M (2004) Analysis of gymnosperm two-AP2-domain-containing genes. Dev genes Environ 214:105–114
Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang PC, Zhu JK (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17:2384–2396
Song X, Li Y, Hou X (2013) Genome-wide analysis of the AP2/ERF transcription factor superfamily in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom 14:573–588
Song X, Wang J, Ma X, Li Y, Lei T, Wang L, Ge W, Guo D, Wang Z, Li C (2016) Origination, expansion, evolutionary trajectory, and expression bias of AP2/ERF superfamily in Brassica napus. Front Plant Sci 7:1186. https://doi.org/10.3389/fpls.2016.01186
Suzuki N, Rivero RM, Shulaev V, Blumwald E, Mittler R (2014) Abiotic and biotic stress combinations. New Phtol 203:32–43
Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z (2017) agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res 45:W122–W129
Tuteja N (2007) Abscisic acid and abiotic stress signaling. Plant Signaling Behav 2:135–138
Wei YL, Li JN, Lu J, Tang ZL, Pu DC, Chai YR (2007) Molecular cloning of Brassica napus TRANSPARENT TESTA 2 gene family encoding potential MYB regulatory proteins of proanthocyanidin biosynthesis. Mol Biol Rep 34(2):105–120
Xiao Q, Zhang F, Nacev BA, Liu JO, Pei D (2010) Protein N-terminal processing: substrate specificity of Escherichia coli and human methionine aminopeptidases. Biochemistry 49:5588–5599
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Zhou J, Tang X, Martin GB (1997) The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J 16:3207–3218
Acknowledgements
The authors are grateful to Shiraz University (Grant No. SH82194-181).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ghorbani, R., Zakipour, Z., Alemzadeh, A. et al. Genome-wide analysis of AP2/ERF transcription factors family in Brassica napus. Physiol Mol Biol Plants 26, 1463–1476 (2020). https://doi.org/10.1007/s12298-020-00832-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-020-00832-z