Abstract
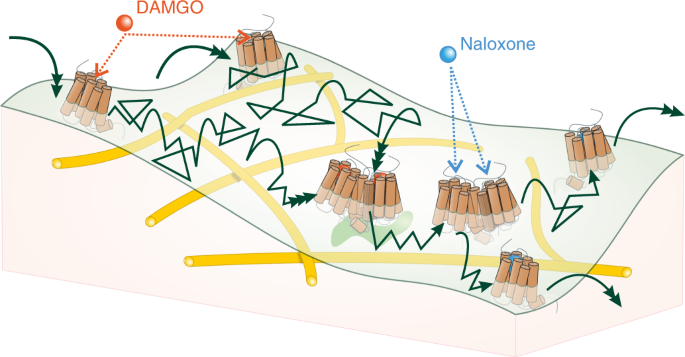
G-protein-coupled receptors (GPCRs) are key signaling proteins that mostly function as monomers, but for several receptors constitutive dimer formation has been described and in some cases is essential for function. Using single-molecule microscopy combined with super-resolution techniques on intact cells, we describe here a dynamic monomer–dimer equilibrium of µ-opioid receptors (µORs), where dimer formation is driven by specific agonists. The agonist DAMGO, but not morphine, induces dimer formation in a process that correlates both temporally and in its agonist- and phosphorylation-dependence with β-arrestin2 binding to the receptors. This dimerization is independent from, but may precede, µOR internalization. These data suggest a new level of GPCR regulation that links dimer formation to specific agonists and their downstream signals.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The authors declare that all the data supporting the findings of this study are available within the paper and its supplementary information files or from the corresponding author upon reasonable request. Source data are provided with this paper.
Code availability
All custom codes used in this study are available through the Github depository mentioned in the related online methods section.
References
Pierce, K. L., Premont, R. T. & Lefkowitz, R. J. Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 3, 639–650 (2002).
Staus, D. et al. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature 535, 448–452 (2016).
Gregorio, G. et al. Single-molecule analysis of ligand efficacy in β2AR-G protein activation. Nature 547, 68–73 (2017).
Kasai, R. S. et al. Full characterization of GPCR monomer–dimer dynamic equilibrium by single molecule imaging. J. Cell Biol. 192, 463–480 (2011).
Calebiro, D. et al. Single-molecule analysis of fluorescently labeled G-protein-coupled receptors reveals complexes with distinct dynamics and organization. Proc. Natl Acad. Sci. USA 2, 743–748 (2013).
Sungkaworn, T. et al. Single-molecule imaging reveals receptor–G protein interactions at cell surface hot spots. Nature 550, 543–547 (2017).
Kniazeff, J., Prézeau, L., Rondard, P., Pin, J.-P. & Goudet, C. Dimers and beyond: the functional puzzles of class C GPCRs. Pharmacol. Ther. 130, 9–25 (2011).
Manglik, A. et al. Crystal structure of the µ-opioid receptor bound to a morphinan antagonist. Nature 485, 321–326 (2012).
Huang, W. et al. Structural insights into µ-opioid receptor activation. Nature 524, 315–321 (2015).
Koehl, A. et al. Structure of the µ-opioid receptor–Gi protein complex. Nature 558, 547–555 (2018).
Sounier, R. et al. Propagation of conformational changes during µ-opioid receptor activation. Nature 524, 375–378 (2015).
Meral, D. et al. Molecular details of dimerization kinetics reveal negligible populations of transient µ-opioid receptor homodimers at physiological concentrations. Sci. Rep. 8, 7705 (2018).
Jordan, B. A. & Devi, L. A. G-protein-coupled receptor heterodimerization modulates receptor function. Nature 399, 697–700 (1999).
Ferré, S. et al. G protein-coupled receptor oligomerization revisited: functional and pharmacological perspectives. Pharmacol. Rev. 66, 413–434 (2014).
Groer, C. et al. An opioid agonist that does not induce µ-opioid receptor–arrestin interactions or receptor internalization. Mol. Pharmacol. 71, 549–557 (2007).
Keppler, A. et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 21, 86–59 (2003).
Sungkaworn, T. et al. High-resolution spatiotemporal analysis of receptor dynamics by single-molecule fluorescence microscopy. J. Vis. Exp. 89, e51784 (2014).
Jaqaman, K. et al. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods 5, 695–702 (2008).
Grimm, J. B. et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250 (2015).
Grimm, J. B. et al. Synthesis of Janelia Fluor HaloTag and SNAP-Tag ligands and their use in cellular imaging experiments. Methods Mol. Biol. 1663, 179–188 (2017).
Greene, J. L. et al. Covalent dimerization of CD28/CTLA-4 and oligomerization of CD80/CD86 regulate T cell costimulatory interactions. J. Biol. Chem. 271, 26762–26771 (1996).
Fries, J. R. et al. Quantitative identification of different single molecules by selective time-resolved confocal fluorescence spectroscopy. J. Phys. Chem. A 102, 6601–6613 (1998).
Huang, P. et al. Functional role of a conserved motif in TM6 of the rat μ opioid receptor: constitutively active and inactive receptors result from substitutions of Thr6.34(279) with Lys and Asp. Biochemistry 40, 13501–13509 (2001).
Miess, E. et al. Multisite phosphorylation is required for sustained interaction with GRKs and arrestins during rapid µ-opioid receptor desensitization. Sci. Signal. 11, 539 (2018).
Heilemann, M. et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 47, 6172–6176 (2008).
Sauer, M. & Heilemann, M. Single-molecule localization microscopy in eukaryotes. Chem. Rev. 117, 7478–7509 (2017).
Dietz, M. S. & Heilemann, M. Optical super-resolution microscopy unravels the molecular composition of functional protein complexes. Nanoscale 11, 17981–17991 (2019).
Karathanasis, C. et al. Molecule counts in localization microscopy with organic fluorophores. ChemPhysChem 18, 942–948 (2017).
Malkusch, S. & Heilemann, M. Extracting quantitative information from single-molecule super-resolution imaging data with LAMA - LocAlization Microscopy Analyzer. Sci. Rep. 6, 34486 (2016).
Fricke, F. et al. One, two or three? Probing the stoichiometry of membrane proteins by single-molecule localization microscopy. Sci. Rep. 5, 14072 (2015).
Grushevskyi, E. et al. Stepwise activation of a class C GPCR begins with millisecond dimer rearrangement. Proc. Natl Acad. Sci. USA 116, 10150–10155 (2019).
Patriarchi, T. et al. Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 360, eaat4422 (2018).
Lohse, M. J., Nuber, S. & Hoffmann, C. Fluorescence/bioluminescence resonance energy transfer techniques to study G-protein-coupled receptor activation and signaling. Pharmacol. Rev. 64, 299–336 (2012).
Yanagawa, M. Single-molecule diffusion-based estimation of ligand effects on G protein-coupled receptors. Sci. Signal. 11, eaao1917 (2018).
Metz, M., Pennock, R., Krapf, D. & Hentges, S. Temporal dependence of shifts in mu opioid receptor mobility at the cell surface after agonist binding observed by single-particle tracking. Sci. Rep. 9, 7297 (2019).
Halls, M. L. et al. Plasma membrane localization of the µ-opioid receptor controls spatiotemporal signaling. Sci. Signal. 9, ra16 (2016).
Jullié, D. et al. A discrete presynaptic vesicle cycle for neuromodulator receptors. Neuron 105, 663–677.e8 (2020).
Gondin, A. B. et al. GRK mediates µ-opioid receptor plasma membrane reorganization. Front. Mol. Neurosci. 12, 104 (2019).
Bhatia, S. et al. Different cell surface oligomeric states of B7-1 and B7-2: implications for signaling. Proc. Natl Acad. Sci. USA 102, 15569–15574 (2005).
Dorsch, S. et al. Analysis of receptor oligomerization by FRAP microscopy. Nat. Methods 6, 225–230 (2009).
Jordan, B. A. et al. Functional interactions between µ opioid and α2A-adrenergic receptors. Mol. Pharmacol. 64, 1317–1324 (2003).
Vilardaga, J. P. et al. Conformational cross-talk between α2A-adrenergic and µ-opioid receptors controls cell signaling. Nat. Chem. Biol. 4, 126–131 (2008).
He, L. et al. Regulation of opioid receptor trafficking and morphine tolerance by receptor oligomerization. Cell 108, 271–282 (2002).
Williams, J. T. et al. Regulation of µ-opioid receptors: desensitization, phosphorylation, internalization, and tolerance. Pharmacol. Rev. 65, 223–254 (2013).
Sommer, M. E. et al. Arrestin–rhodopsin binding stoichiometry in isolated rod outer segment membranes depends on the percentage of activated receptors. J. Biol. Chem. 286, 7359–7369 (2011).
Beyrière, F. et al. Formation and decay of the arrestin·rhodopsin complex in native disc membranes. J. Biol. Chem. 290, 12919–12928 (2015).
Lohse, M. J. et al. in Arrestins-Pharmacology and Therapeutic Potential (ed. Gurevich, V. V.) 15–56 (Springer, 2014).
van Unen, J. et al. A new generation of FRET sensors for robust measurement of Gαi1, Gαi2 and Gαi3 activation kinetics in single cells. PLoS ONE 11, e0146789 (2016).
Ovesný, M., Kˇrížek, P., Borkovec, J., Švindrych, Z. & Hagen, G. M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30, 2389–2390 (2014).
Wolter, S. et al. Real-time computation of subdiffraction-resolution fluorescence images. J. Microsc. 237, 12–22 (2010).
Hummer, G., Fricke, F. & Heilemann, M. Model-independent counting of molecules in single-molecule localization microscopy. Mol. Biol. Cell 27, 3637–3644 (2016).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Dupont, A., Stirnnagel, K., Lindemann, D. & Lamb, D. Tracking image correlation: combining single-particle tracking and image correlation. Biophys. J. 104, 2373–2382 (2013).
Hoffmann, M., Fröhner, C. & Noé, F. ReaDDy 2: fast and flexible software framework for interacting-particle reaction dynamics. PLoS Comput. Biol. 15, e1006830 (2019).
Serfling, R. et al. Quantitative single-residue bioorthogonal labeling of G protein-coupled receptors in live cells. ACS Chem. Biol. 14, 1141–1149 (2019).
Trullo, A., Corti, V., Arza, E., Caiolfa, V. R. & Zamai, M. Application limits and data correction in number of molecules and brightness analysis. Microsc. Res. Tech. 76, 1135–1146 (2013).
Isbilir, A. et al. Visualization of class A GPCR oligomerization by image-based fluorescence fluctuation spectroscopy. Preprint at BioRxiv https://doi.org/10.1101/240903 (2017).
Acknowledgements
We thank L. Lavis (Janelia Research Campus) for providing sufficient amounts of halo-tagged JF-646 dye. We further thank U. Zabel and M. Frank for molecular cloning. This work was supported by the Deutsche Forschungsgemeinschaft, TR166 (to M.J.L.), the National Institutes of Health grant DA038882 (to M.J.L.), the cluster of excellence MATH+ (to C.S. and M.J.L.) and the Elitenetzwerk Bayern, Receptor Dynamics program (to M.J.L.). B.O. received funding by the EU Horizon 2020 research and innovation program, CORBEL, grant no. 654248.
Author information
Authors and Affiliations
Contributions
M.J.L., J.M., T.S., V.S., C.K., M.H., A.B., P.A. and C.S. conceived the experiments. J.M., A.I., C.K., V.S. and P.A. conducted the experiments. J.M., B.O., T.S., E.O.G. and P.A. analyzed the results. B.O. programmed the Polytracker code. M.J.L. and J.M. wrote the manuscript. M.J.L. initiated the project. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Fitting of individual PSFs, single molecule tracking and extended diffusion analyses.
a, Diffraction limited single molecule image (left) of SNAP-tagged receptor labeled with SNAP-Surface 549® Dye. Fitting of individual PSFs (right) allows a localization with sub-pixel precision of approx. 25 nm. Camera pixels are depicted as cyan grid (pixel-size 107 nm). b, Tracks of single molecule movies classified by u-Track based on their confinement radius and diffusion coefficient. c, Fraction of tracks undergoing actin-crossing events at corresponding time-points after receptor activation with 10 µM DAMGO. Shown are mean ± SEM (n = 5 cells for basal and 1 min, n = 4 for 5 min, n = 3 for 10 min, and n = 2 for 15 min). P values were determined by one-way ANOVA and Tukey’s multiple comparison test. d, Fractions of the diffusion classes shown in Fig. 1 over time. [(a, b) are representative images. Experiments in (c) are from n = 19 cells and 3 independent experimental days. (d) statistics are based on the experiments shown in Fig. 1].
Extended Data Fig. 2 µOR compartmentalization and diffusion classes.
a, Diffraction limited image of representative live-cell with dual-color acquisition of µORs (magenta) and actin (green)-(See also Supplementary Video 1). Note that the receptor spots in the bottom right corner belong to another cell that does not express LifeAct. b, Super-resolved image of the cell in (a) showing that the µOR is compartmentalized by actin fibers. The track-color corresponds with the type of diffusion (magenta = directed diffusion, blue= confined diffusion, cyan= Brownian motion and brown= immobile) (See also Supplementary Video 2) [Shown images are representative of five independent experiments].
Extended Data Fig. 3 Density limitation of single molecule tracking and simulation of random co-localization events.
a, Amount of dimerization events occurring for the basal wild-type µOR. The linear fit (magenta) shows the expression range up to which strict single molecule analysis can be conducted without bias due to overlapping of PSFs. b, Representative image of simulated movies comprising the same distribution of diffusion classes and diffusion- coefficients as real movies. Particles undergoing confined and directed diffusion were modelled using different potential fields as described in the methods section. [Data in (a) is fitted from n = 20 cells imaged in three independent experiments. Numbers and statistics on the simulations are provided in methods section.].
Extended Data Fig. 4 Validation of receptor dimerization with brightness analyses and FRET at higher expression levels.
a, Temporal molecular brightness measurements, normalized to oligomeric state, for corresponding C-terminally tagged YFP constructs confirm that the β1-AR (normalized brightness= 0.965 ± 0.054, n = 47) and µOR (normalized brightness= 1.087 ± 0.051, n = 55) are largely monomeric. In contrast, CD28 results in an almost perfectly doubled brightness (brightness = 1.929 ± 0.111, n = 19). These experiments show that dimerization is not influenced by the N-terminal SNAP-tag used for single molecule studies. b, SpIDA brightness measurements with SNAP-tagged receptor constructs show that results also agree when SNAP-constructs are used in brightness measurements (normalized brightness values: SNAP-β1AR = 0.949 ± 0.118, n = 10; SNAP-µOR = 0.975 ± 0.081, n = 19; SNAP-µOR + 10 µM morphine = 0.987 ± 0.067, n = 16; SNAP-µOR + 10 µM DAMGO = 1.843 ± 0.121, n = 12; µOR (T279K) mutant 1.755 ± 0.076, n = 20; SNAP-CD28 = 2.1 ± 0.109, n = 19). Receptor expression levels used in these experiments are ≈ 40 receptors per µm2. Brightness values in are mean ± SEM and n = number of cells. The box shows IQR and whiskers are SD. P values were determined by one-way ANOVA and Tukey’s multiple comparison test. c, Acceptor photobleaching experiments on a confocal microscope with dual-color labeled SNAP-constructs also confirm the monomeric nature of β1AR and the µOR by linearly increasing FRET-efficiencies at high expression levels. In contrast, the dimer control SNAP-CD28 shows a steep increase and quick saturation of the measured FRET-efficiencies. d, The inactive T279D mutant of the µOR does not increase significantly the FRET- efficiency between receptor protomers. In contrast the T279K mutant shows significantly higher FRET-efficiencies over increasing expression. Due to the constitutive activity of this mutant, expression levels higher than depicted could not be achieved. Curves for SNAP-β1AR (blue) and SNAP-µOR-wt (black) are taken from panel c. Graphs show pooled experiments repeated independently n = 3 times on 3 different experimental days. Each datapoint represents a value from one individual cell.
Extended Data Fig. 5 Characterization of µOR-mutants shows high basal activity of the T279K mutant and impaired activation of the T279D mutant.
a, 24-hour pre-treatment with 100 ng/mL PTX abolishes high-affinity binding (≤ 1 %) due to Gαi inactivation. Wt- data from Supplementary Fig. 1 (n = 3 independent experiments). The PTX curve is a representative one of 3 independent experiments with similar results, datapoints of the curve are mean ± SD. b, Fraction of high- affinity binding is dramatically increased for the constitutively active T279K mutant (64.4 ± 1.6 %). High- affinity binding is abolished in the T279D mutant (≤ 1 %), comparable to PTX treated wild-type receptor. Data are mean ± SEM from n = 3 independent experiments of each condition. P values were determined by one-way ANOVA and Tukey’s multiple comparison test. c, For the T279K mutant G protein activation occurs already without DAMGO stimulation, leading to a full response of the Gi- FRET sensor in the basal state. The response of the T279D mutant is right-shifted to the wild-type (pEC50-wt= 9.2 ± 0.1 vs. pEC50-T279D = 6.6 ± 0.1). Data are mean ± SEM from n = 4 independent experiments of each variant. d, Measurement of basal receptor activity and its mutants, based on Gi-mediated cAMP decrease, after receptor activation with DAMGO. Cells were pre-stimulated with 1 µM forskolin to slightly elevate cAMP levels. The pEC50 of the constitutively active mutant (10.1 ± 0.1) does not significantly differ from µOR-wild-type (9.9 ± 0.4). Due to the high basal activity of the T279K mutant the amplitude of additional Gi activation via DAMGO is dramatically reduced (20.8 ± 1.7 %), compared to the wild-type receptor (100 %). The inactive T279D mutant shows a pEC50 right-shifted by 2–3 log-units (7.4 ± 0.1) while the amplitude is slightly but not significantly higher than the one of the wild-type receptor (103 ± 2 %). Data from n = 4 independent experiments. Numbers are means and errors are given as SEM.
Extended Data Fig. 6 Effects of naloxone/DAMGO competition on µOR dimerization measured via intensity distribution.
a,b, Representative intensity distribution analyses based on TIRF images of wild-type µOR at different time points, with naloxone and DAMGO (10 µM each) added as indicated: naloxone first (a) or DAMGO first (b). When naloxone is applied first, the µOR intensity distribution remains monomeric even after subsequent addition of DAMGO, indicating that naloxone prevents activation-dependent dimerization. When DAMGO is given first, the µOR intensity distribution indicates dimerization after 15 min, and addition of naloxone results in disruption of dimers as indicated by a progressive return to a rather monomeric distribution over the next 15 min. [Data were generated as in Fig. 2 and are from 314 (a) or 470 (b) detected PSFs from 7 (a) or 6 (b) cells. Experiments were repeated independently 3 times with similar results].
Extended Data Fig. 7 µOR internalization after receptor activation is spatially and temporally separated from receptor dimerization.
a, Dual- color single molecule TIRF images illustrate that the unstimulated wild-type receptor as well as the T279K mutant show negligible co-localization with clathrin. Using a steeper TIRF- angle and thereby increasing the penetration depth of the laser shows receptor co-localizations of higher order intensities (up to ten fluorophores per spot) and co-localization with clathrin underneath the plasma- membrane after 20 min, and more strongly after 45 min, of stimulation by 10 µM DAMGO, illustrating µOR internalization. b, Comparative images of µOR-wild type and T279K mutant show that the diffusion pattern of the T279K mutant is also receptor-typical (in contrast to pits or endosomes). c, Concentration-response curve of a BRET-based internalization assay showing that DAMGO is leading to receptor internalization with a pIC50 of 6.3 ± 0.1. In contrast, morphine is not leading to receptor internalization at any concentration. d, Internalization kinetics after stimulation with 10 µM DAMGO using the assay shown in (c) reveals a τ- value of 11.7 ± 0.3 min. [Images (a, b) are each representatives of five independent experiments. Datapoints in (c) and (d) are means and errors are given as SEM. Data shown in (c) are n = 3 independent experiments and (d) are n = 4 individual kinetic measurements].
Extended Data Fig. 8 Quantitative dSTORM microscopy of monomeric and dimeric controls.
Super-resolved and brightfield (inset) images as well as histograms of the number of blinking events for Alexa Fluor 647® conjugated to the SNAP-tag of the respective protein are shown. The blinking histogram was fitted with theoretical model functions reporting on monomer and dimer populations (shown in pie charts). SNAP-β1AR (a) and SNAP-CD28 (b) serve as calibration standards for monomeric and dimeric proteins, which allows robust extraction of analysis parameters. Analysis reveals 100 ± 3 % monomers for SNAP-β1AR and SNAP-µOR, 100 ± 3 % dimers for SNAP-CD28. [Data are mean ± SD. In total 714 spots for SNAP-β1AR and 661 spots for SNAP-CD28 were analyzed. For both controls, n = 12 cells from three independent experiments were analyzed].
Extended Data Fig. 9 Oligomeric state characterization of the phosphorylation deficient constitutively active T279K mutant and verification of effectors on the wild-type µOR.
a, Intensity histograms of the dimeric T279K mutant and its phosphorylation-deficient modification. The phosphorylation-deficient modified mutant shows a monomeric distribution of intensities. b, Bleaching experiments over 400 frames (each 40 ms) and high laser-power reveal an almost exactly doubled half-life time for the T279K mutant (8.9 ± 0.3 s) in contrast to its phosphorylation-deficient modification (4.5 ± 0.2 s). c, Oligomerization states of wild-type µOR, its phosphorylation-deficient modification (pd) as well as wild-type µOR treated with Pitstop2 (Ps2) before and after activation with 10 µM DAMGO. The phosphorylation-deficient modification results in a monomeric distribution of intensities. Pitstop2 had within our measurement time window no significant effect on the basal monomeric state of the µOR, nor on the DAMGO-induced formation of dimers. d, Individual datapoints and significance of the measurements shown in (c). Whiskers show SD, the box represents IQR of the data and the line indicates the median. [Data in (a) were generated from 347 (T279K-pd) and 287 (T279K) detected PSFs from 5 different cells tested on 3 experimental days. (b) shows a representative bleaching trace over 400 frames extracted from datasets acquired in (a). Data in c and d are from n = 8, 5, 5, 5, 4, and 6 different cells for “wt”, “wt+DAMGO”, “wt-pd”, “wt-pd+DAMGO”, “wt+Ps2” and “wt+Ps2+DAMGO”, respectively. Cells of each variant are from three independent experiments. One Datapoint in (d) corresponds to one PSF- intensity. P values were determined by one-way ANOVA and Tukey’s multiple comparison test].
Supplementary information
Supplementary Information
Supplementary Figs. 1–3, Table 1 and Note.
Supplementary Video 1
Single-molecule imaging of individual µ-opioid receptors among actin fibers at the surface of living cells. Diffraction limited dual-color movie of actin (green) and µOR (magenta), showing that actin fibers contribute to receptor compartmentalization.
Supplementary Video 2
Actin imaging and receptor tracking with sub-pixel precision reveals diffusion behavior. Shown is a sub-pixel resolution inset of the cell shown in Supplementary Video 1, overlaid with particle tracking of µORs and its corresponding diffusion classification, to further elucidate compartmentalization of the µOR. The diffusive behavior of the tracks is classified in immobile (brown), confined (blue), Brownian (cyan) and directed (magenta). For actin the color code ‘yellow’ of Fiji ImageJ was applied.
Supplementary Video 3
Single-molecule imaging of individual receptors among clathrin-coated pits at the surface of living cells. Diffraction-limited dual-color movie of clathrin-coated pits (imaged using σ2-AP2-mNeonGreen) (green) and the constitutively active T279K mutant of the µOR (magenta). Images were acquired after SNAP labeling, showing that dimers of this receptor mutant are initially not accumulated in clathrin-coated pits.
Supplementary Video 4
SNAP-Surface 549 dye allows robust single-molecule imaging with fast acquisition. Single-color movie with 1,000 frames to illustrate that the used dye shows enough photostability in fast image acquisitions as required for single-particle tracking. Note that just up to the 400 first frames are used for image analysis in this manuscript.
Supplementary Video 5
Labeling with SNAP-Surface 549 dye enables high signal to noise imaging. Single-color movie with 1,000 frames to further illustrate the photostability and bleaching behavior of the used dye in image acquisitions with high laser power. Such settings are used for intensity analysis (just the first frame is used in this case) or bleaching kinetics measurements.
Supplementary Data 1
Source Data for Supplementary Figs. 1–3
Source data
Source Data Fig. 1
Source Data of the shown panels.
Source Data Fig. 2
Source Data of the shown panels.
Source Data Fig. 3
Source Data of the shown panels.
Source Data Fig. 5
Source Data of the shown panels.
Source Data Fig. 6
Source Data of the shown panels.
Source Data Extended Data Fig. 1
Source Data of the shown panels.
Source Data Extended Data Fig. 4
Source Data of the shown panels.
Source Data Extended Data Fig. 5
Source Data of the shown panels.
Source Data Extended Data Fig. 6
Source Data of the shown panels.
Source Data Extended Data Fig. 7
Source Data of the shown panels.
Source Data Extended Data Fig. 8
Source Data of the shown panels.
Source Data Extended Data Fig. 9
Source Data of the shown panels.
Rights and permissions
About this article
Cite this article
Möller, J., Isbilir, A., Sungkaworn, T. et al. Single-molecule analysis reveals agonist-specific dimer formation of µ-opioid receptors. Nat Chem Biol 16, 946–954 (2020). https://doi.org/10.1038/s41589-020-0566-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-0566-1
This article is cited by
-
The chemokine receptor CCR5: multi-faceted hook for HIV-1
Retrovirology (2024)
-
Computational drug development for membrane protein targets
Nature Biotechnology (2024)
-
Quantitative determination of fluorescence labeling implemented in cell cultures
BMC Biology (2023)
-
Dual-color DNA-PAINT single-particle tracking enables extended studies of membrane protein interactions
Nature Communications (2023)
-
Atomic force microscopy-single-molecule force spectroscopy unveils GPCR cell surface architecture
Communications Biology (2022)