Abstract
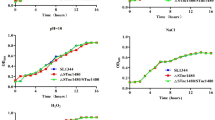
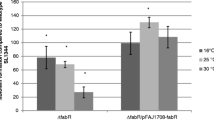
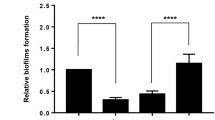
In this study, we aimed at identifying the regulatory role of marT gene, known as the regulator of misL, on 15 different biofilm-related genes in S. Typhimurium 14028 strain. We also tested the strains for their ability to form biofilm and determined the adherence characteristics of the wild type and the mutant strains of the organism on Caco-2 and HEp-2 cells. For comparative analyses of the candidate genes, individual gene mutations were created via antibiotic gene cassette insertion into each gene of interest. marT gene was cloned behind an arabinose inducible BAD promoter in order to control marT expression. This recombinant plasmid was transfer into each of the 15 mutant strains to investigate the level of expression of each single gene in the presence and absence of marT induction. Besides determination of variations in biofilm formation by each mutant strain, the attachment characteristics of them onto Caco-2 and HEp-2 cell lines were also reported. As a result of attachments experiments on polystyrene surfaces, it was determined that the biofilm production capacity of each mutant strain decreased in a statistically significant manner (p < 0.05). QRT-PCR trials indicated that the marT gene regulates the expression of 14 genes, namely fimA, fimD, fimF, fimH, stjB, stjC, csgA, csgD, ompC, sthB, sthE, rmbA, fliZ and yaiC, in a positive manner. QRT-PCR studies were also revealed that the MarT protein positively regulates its own promoter. When the adherence characteristics of the mutant strains and the wild-type were investigated by using Caco-2 and HEp-2 cells, it was determined that the single gene mutations did have no effect on bacterial adhesion. In view of our mutational analyses and biofilm formation studies, it was concluded that fliZ, ompC, rmbA, stjB and stjC genes are related with biofilm formation in Salmonella, besides other cellular functions of them. Taken together, our data suggested that the regulatory role of MarT protein is not only restricted to the regulation of misL gene expression, but it rather acts as a general regulator on the biofilm-related genes in Salmonella.





Similar content being viewed by others
References
Pui CF, Wong WC, Chai LC, Nillian E, Ghazali FM, Cheah YK, Nakaguchi Y, Nishibuchi M, Radu S (2011) Simultaneous detection of Salmonella spp., Salmonella Typhi and Salmonella Typhimurium in sliced fruits using multiplex PCR. Food Control 22:337–342
Stepanovic S, Irkovi I, Ranin L, Svabi-Vlahovi M (2004) Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett Appl Microbiol 38:428–432
Barnhart MM, Chapman MR (2006) Curli biogenesis and function. Annu Rev Microbiol 60:131–147
Ledeboer NA, Jones BD (2005) Exopolysaccharide sugars contribute to biofilm formation by Salmonella enterica serovar Typhimurium on HEp–2 cells and chicken intestinal epithelium. J Bacteriol 187:3214–3226
Ledeboer NA, Frye JG, McClelland M, Jones BD (2006) Salmonella enterica serovar Typhimurium requires the Lpf, Pef, and Tafi fimbriae for biofilm formation on HEp-2 tissue culture cells and chicken intestinal epithelium. Infect Immun 74:3156–3169
Boddicker JD, Ledeboer NA, Jagnow J, Jones BD, Clegg S (2002) Differential binding to and biofilm formation on, HEp-2 cell by Salmonella enterica serovar Typhimurium is dependent upon allelic variation in the fimH gene of the fim gene cluster. Mol Microbiol 45:1255–1265
Da Re S, Ghigo J (2006) A csgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J Bacteriol 188:3073–3087
Ghigo JM (2001) Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442–445
Reisner A, Haagensen JAJ, Schembri MA, Zechner EL, Molin S (2003) Development and maturation of Escherichia coli K-12 Biofilms. Mol Microbiol 48:933–946
Tükel Ç, Akçelik M, de Jong MF, Şimşek Ö, Tsolis RM, Baumler AJ (2007) MarT activates expression of the MisL autotransporter protein of Salmonella enterica serotype Typhimurium. J Bacteriol 189:3922–3926
Akçelik N (2011) In vitro production and identification of protein-protein interactions of MisL autotransporter protein of Salmonella Typhimurium. PhD Thesis, Faculty of Science, Ankara University
Yener İlçe B, Akçelik N (2015) The effect of marT gene on biofilm production of Salmonella Typhimurium. Turk J Biol 39:722–731
Vestby LK, Møretrø T, Langsrud S, Heir E, Nesse LL (2009) Biofilm forming abilities of Salmonella are correlated with persistence in fish meal- and feed factories. BMC Vet Res 5:20
Hancox LS, Yeh KS, Clegg S (1997) Construction and characterisation of type 1 non-fimbriate and non-adhesive mutants of Salmonella Typhimurium. FEMS Immunol Med Microbiol 19:89–296
Zeiner SA, Dwyer BE, Clegg S (2012) fimA, fimF, fimH are necessary for assembly of type 1 fimbriae on Salmonella enterica serovar Typhimurium. Infect Immun 80:3289–3296
Akkoç N, Özden B, Tan BG, Akçelik M (2009) The role of stj fimbrial operon in the intestinal persistence of Salmonella Typhimurium in mice. Biologia 64:859–863
Yüksel D, Akçelik N, Akçelik M (2012) Transcriptional regulation of stj fimbrial operon in Salmonella Typhimurium. Ankara Üniv Vet Fak Derg 59:65–70
Negm RS, Pistole TG (1999) The Porin ompC of Salmonella Typhimurium mediates adherence to macrophages. Can J Microbiol 45:658–669
Townsend SM, Kramer NE, Edvards R, Baker S, Hamlin N, Simmonds M, Stevens K, Maloy S, Parkhill J, Dougan G, Baumler AJ (2001) Salmonella enterica serovar Typhi possesses a unique repertoire of fimbrial gene sequences. Infect Immun 69:2894–2901
Latasa C, Roux A, Toledo-Arana A, Ghigo JM, Gamazo C, Penadés JR, Lasa I (2005) BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol Microbiol 58:1322–1339
Liu Z, Niu H, Wu S, Huang R (2014) CsgD Regulatory network in bacterial trait-altering biofilm formation. Emerg Microbes Infect 3(1):e1
Cowles KN, Willis DK, Engel TN, Jones JB, Barak JD (2016) Diguanylate cyclases adrA and SRM1987 regulate Salmonella enterica exopolysaccharide production during plant colonization in an environment-dependent manner. Appl Environ Microbiol 82:1237–1248
Gottesman S (1984) Bacterial regulation: global regulatory networks. Ann Rev Genet 18:415–441
Perez-Rueda E, Collardo-Vides J (2000) The reportaire of DNA-binding transcriptional regulators in Escherichia coli K 12. Nucleic Acid Res 28:1838–1847
Guiterrez-Rois RM, Rosernblueth DA, Loza JA, Huerta AM, Glasner JD (2003) Regulatory network of Escherichia coli: consistency between literature knowledge and microarray profiles. Genome Res 13:2435–2443
Ishihama A (2010) Prokaryotic genome regulation: multifactor promoters, multitarget regulators and hierarchic networks. FEMS Microbiol Rev 34:628–645
Morgan E, Campbell JD, Rowe SC, Bispham J, Stevens MP, Bowen AJ, Barrow PA, Maskell DJ, Wallis TS (2004) Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol Microbiol 54:994–1010
Crawford RW, Rosales-Reyes R, Ramirez-Aguilar Mde L, Chapa-Azuela O, Alpuche- Aranda C, Gunn JS (2010) Gallstones play a significant role in Salmonella spp.gallbladder colonization and carriage. Proc Natl Acad Sci USA 107:4353–4358
Gonzales-Escobedo G, Gunn JS (2013) Gallbladder epithelium as a niche for chronic Salmonella carriage. Infect Immun 81(8):2920–2930
El Hag M, Feng Z, Su Y, Wang X, Yassin A, Chen S, Peng D, Liu X (2017) Contribution of the csgA and bcsA genes to Salmonella enterica serovar Pullorum biofilm formation and virulence. Avian Pathol 46:541–547
MacKenzie KD, Palmer MB, Köster WL, White AP (2017) Examining the link between biofilm formation and the ability of pathogenic Salmonella strains to colonize multiple host species. Front Vet Sci 4:138. https://doi.org/10.3389/fvets.2017.00138
Blanc-Potard AB, Solomon F, Kayser J, Groisman EA (1999) The SPI-3 patgogenicity island of Salmonella enterica. J Bacteriol 181:998–1004
Guttenplan SP, Kearns DP (2013) Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev 37:849–871
De Masi L, Yue M, Hu C, Takov AV, Rankin SC, Schifferli DM (2017) Cooperation of adhesin allelles in Salmonella-host tropism. mSphere 2:e00066–e117
Berne C, Ducret A, Hardy GG, Brun YV (2015) Adhesins involved in attachment to abiotic surfaces by Gram negative bacteria. Microbiol Spectr. https://doi.org/10.1128/microbiolspec.MB-0018-2015
Acknowledgements
This work was supported by the grant 114Z871 from The Scientific and Technological Research Council of Turkey (TÜBİTAK). We would like to thank to Prof. Dr. İhsan Gürsel, Prof. Dr. Erkan Yılmaz, Prof. Dr. Sreeparna Banerjee and Professor Michael McClelland for their contributions.
Author information
Authors and Affiliations
Contributions
ZE and BCY performed the experimental analyses. MA and GÖ guided the experiments and wrote the original draft. NA established the main idea of the manuscript, supervised the study, and edited the original draft.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2020_5573_MOESM2_ESM.png
Supplemental Fig. 1 PCR results verifying the entry of the kanamycin cassette into the targetgenes. The presence of 193bp amplicone was provedn the presence of the antibiotic cassette inthe target gene. (PNG 711 kb)
Rights and permissions
About this article
Cite this article
Eran, Z., Akçelik, M., Yazıcı, B.C. et al. Regulation of biofilm formation by marT in Salmonella Typhimurium. Mol Biol Rep 47, 5041–5050 (2020). https://doi.org/10.1007/s11033-020-05573-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05573-6