Abstract
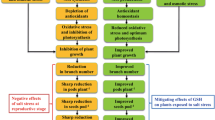
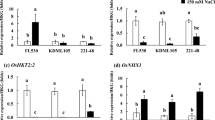
Soil salinity is an environmental stress severely impacting on rice grain yield. However, limited information is available on how salinity affects expression levels of genes determining grain yield. In this study, we investigated agronomic traits associated with grain yield among three japonica rice cultivars grown either under moderate salinity with an electrical conductivity of 4 dS/m or under non-saline conditions in a paddy field in Dongying, Shandong, China. Moderate salinity affected rice yield predominantly by reducing grain number and grain filling. We compared expression levels of genes determining grain number in young panicles (0.5–1 cm in length) of plants grown under salinity or non-saline conditions. Transcription of Lax panicle 1, Lax panicle 2, Ideal plant architecture (IPA1), Dense and erect panicle 1, Tawawa 1, and OsMADS1 was significantly repressed, whereas that of Cytokinin oxidase/dehydrogenase, Grain number per panicle 1, and Narrow leaf 1 was not significantly influenced by salinity. OsmicroRNA156, the posttranscriptional regulator of IPA1, was induced by salinity in the panicle and seedlings and complemented the expression patterns of IPA1. This result implied that the OsmicroRNA156–IPA1 pathway was involved in rice salinity responses. The grain filling-associated genes Grain incomplete filling 1, Grain incomplete filling 2, and Nuclear factor Y were down-regulated by salinity in the spikelet at 5 or 10 days after fertilization, which contributed to the salinity-triggered reduction in grain weight. These findings suggest some targets that may be utilized to improve the grain yield under salinity stress conditions via breeding and gene-pyramiding approaches.







Similar content being viewed by others
References
Abdullah Z, Khan MA, Flowers T (2001) Causes of sterility in seed set of rice under salinity stress. J Agron Crop Sci 187:25–32
An LS (2012) Water-salt characters of groundwater and ecological effects in the Yellow River Delta. Dissertation, Ocean University of China
Aslam M, Qureshi RH, Ahmed N (1993) A rapid screening technique for salt tolerance in rice (Oryza sativa L.). Plant Soil 150:99–107
Fujita D, Trijatmiko KR, Tagle AG, Sapasap MV, Koide Y, Sasaki K, Tsakirpaloglou N, Gannaban RB, Nishimura T, Yanagihara S, Fukuta Y, Koshiba T, Slamet-Loedin IH, Ishimaru T, Kobayashi N (2013) NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars. Proc Natl Acad Sci USA 110:20431–20436
Hakim M, Juraimi AS, Hanafi M, Ali E, Ismail MR, Selamat A, Karim SR (2014) Effect of salt stress on morpho-physiology, vegetative growth and yield of rice. J Environ Biol 35:317–326
Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41(4):494–497
Huang Y, Zhao S, Fu Y, Sun H, Ma X, Tan L, Liu F, Sun X, Sun H, Gu P (2018) Variation in the regulatory region of FZP causes increases in secondary inflorescence branching and grain yield in rice domestication. Plant J 96:716–733
Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345:646–651
Jeon JS, Jang S, Lee S, Nam J, Kim C, Lee SH, Chung YY, Kim SR, Lee YH, Cho YG, An G (2000) leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12:871–884
Jeong DH, Park S, Zhai J, Gurazada SG, De Paoli E, Meyers BC, Green PJ (2011) Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23:4185–4207
Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, Qian Q, Li J (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42:541–544
Komatsu M, Maekawa M, Shimamoto K, Kyozuka J (2001) The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev Biol 231:364–373
Komatsu M, Chujo A, Nagato Y, Shimamoto K, Kyozuka J (2003) FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130:3841–3850
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Li N, Li Y (2016) Signaling pathways of seed size control in plants. Curr Opin Plant Biol 33:23–32
Li X, Qian Q, Fu Z, Wang Y, Xiong G, Zeng D, Wang X, Liu X, Teng S, Hiroshi F (2003) Control of tillering in rice. Nature 422:618
Lu Z, Yu H, Xiong G, Wang J, Jiao Y, Liu G, Jing Y, Meng X, Hu X, Qian Q (2013) Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 25:3743–3759
Lu Z, Shao G, Xiong J, Jiao Y, Wang J, Liu G, Meng X, Liang Y, Xiong G, Wang Y (2015) MONOCULM 3, an ortholog of WUSCHEL in rice, is required for tiller bud formation. J Genet Genomics 42:71–78
Lutts S, Kinet JM, Bouharmont J (1995) Changes in plant response to NaCl during development of rice (Oryza sativa L.) varieties differing in salinity resistance. J Exp Bot 46:1843–1852
Maas EV, Grattan S (1999) Crop yields as affected by salinity. Agronomy 38:55–110
Mittler R, Blumwald E (2010) Genetic engineering for modern agriculture: challenges and perspectives. Ann Rev Plant Biol 61:443–462
Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42:545–549
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Ann Rev Plant Biol 59:651–681
Panda D, Ghosh D, Kar M (2013) Effect of salt stress on the pigment content and yield of different rice (Oryza sativa L.) Ggenotypes. Int J Bio-Res Stress Manag 4:431
Puteh A, Mondal M (2014) Salinity stress during booting and heading stages affects yield in rice. Life Sci J 11:223–226
Puteh AB, Mondal MMA (2016) Salinity stress effect on ion uptake and yield attributes in rice. Life Sci J 13:64–67
Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37:1141–1146
Sasaki T, Ashikari M (2018) Rice genomics, genetics and breeding. Springer, Singapore, pp 191–206
Summart J, Thanonkeo P, Panichajakul S, Prathepha P, McManus MT (2010) Effect of salt stress on growth, inorganic ion and proline accumulation in Thai aromatic rice, Khao Dawk Mali 105, callus culture. Afr J Biotechnol 9:142–152
Tabuchi H, Zhang Y, Hattori S, Omae M, Shimizu-Sato S, Oikawa T, Qian Q, Nishimura M, Kitano H, Xie H (2011) LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 23:3276–3287
Tanaka W, Ohmori Y, Ushijima T, Matsusaka H, Matsushita T, Kumamaru T, Kawano S, Hirano HY (2015) Axillary meristem formation in rice requires the WUSCHEL Ortholog TILLERS ABSENT1. Plant Cell 27:1173–1184
Thitisaksakul M, Tananuwong K, Shoemaker CF, Chun A, O-u-m T, Labavitch JM, Beckles DM (2015) Effects of timing and severity of salinity stress on rice (Oryza sativa L.) yield, grain composition, and starch functionality. J Agric Food Chem 63:2296–2304
Tilman D, Balzer C, Hill J, Befort BL (2011) Global food demand and the sustainable intensification of agriculture. Proc Natl Acad Sci USA 108:20260–20264
Tripathi AK, Pareek A, Sopory SK, Singla-Pareek SL (2012) Narrowing down the targets for yield improvement in rice under normal and abiotic stress conditions via expression profiling of yield-related genes. Rice 5:37
Wang B, Wang H (2017) IPA1: a new “green revolution” gene? Mol Plant 10:779–781
Wang E, Wang J, Zhu X, Hao W, Wang L, Li Q, Zhang L, He W, Lu B, Lin H (2008) Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet 40:1370–1374
Wei X, Jiao G, Lin H, Sheng Z, Shao G, Xie L, Tang S, Xu Q, Hu P (2017) GRAIN INCOMPLETE FILLING 2 regulates grain filling and starch synthesis during rice caryopsis development. J Integr Plant Biol 59:134–153
Xie K, Wu C, Xiong L (2006) Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol 142:280–293
Xing Y, Zhang Q (2010) Genetic and molecular bases of rice yield. Ann Rev Plant Biol 61:421–442
Xu J, Zhang X, Xue H (2016) Rice aleurone layer specific OsNF-YB1 regulates grain filling and endosperm development by interacting with an ERF transcription factor. J Exp Bot 67:6399–6411
Yoshida A, Sasao M, Yasuno N, Takagi K, Daimon Y, Chen R, Yamazaki R, Tokunaga H, Kitaguchi Y, Sato Y (2013) TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc Natl Acad Sci USA 110:767–772
Zeng L, Lesch SM, Grieve CM (2003) Rice growth and yield respond to changes in water depth and salinity stress. Agric Water Manag 59:67–75
Zhang G-H, Li S-Y, Wang L, Ye W-J, Zeng D-L, Rao Y-C, Peng Y-L, Hu J, Yang Y-L, Xu J (2014) LSCHL4 from japonica cultivar, which is allelic to NAL1, increases yield of indica super rice 93–11. Mol Plant 7:1350–1364
Acknowledgements
This study was supported by the by the National Key Research and Development Program of China (2016YFD0100600), the Natural Science Foundation of Shandong Province (ZR2018ZC08N2), Agricultural Variety Improvement Project of Shandong Province (2019LZGC003) and Young Talents Training Program of Shandong Academy of Agricultural Sciences, the Science and Technology Innovation Project of Shandong Academy of Agricultural Sciences (CXGC2018E16). We thank Robert McKenzie, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Funding
This study was supported by the by the National Key Research and Development Program of China (2016YFD0100600), the Natural Science Foundation of Shandong Province (ZR2018ZC08N2), Agricultural Variety Improvement Project of Shandong Province (2019LZGC003) and Young Talents Training Program of Shandong Academy of Agricultural Sciences, the Science and Technology Innovation Project of Shandong Academy of Agricultural Sciences (CXGC2018E16).
Author information
Authors and Affiliations
Contributions
XX designed the experiments. CZ and ZZ performed the molecular experiments. GZ and YP planted materials and completed phenotypic analysis. WL analyzed the data. CZ and XX wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical Approval
The authors declare that the experiments comply with the laws of China.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zheng, C., Zhou, G., Zhang, Z. et al. Moderate Salinity Stress Reduces Rice Grain Yield by Influencing Expression of Grain Number- and Grain Filling-Associated Genes. J Plant Growth Regul 40, 1111–1120 (2021). https://doi.org/10.1007/s00344-020-10168-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-020-10168-3