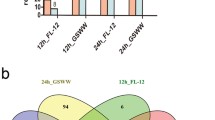
Abstract
Porcine epidemic diarrhea (PED) is an acute enteric disease caused by porcine epidemic diarrhea virus (PEDV). In China, variant PEDV causes severe watery diarrhea, vomiting, and dehydration in piglets, leading to very high morbidity and mortality. However, the pathogenesis of PEDV is still not fully understood. In our study, we analyzed the long noncoding RNA (lncRNA) and mRNA expression profiles of PEDV GDgh16 in infected Vero cells at 60 h postinfection. A total of 61,790 annotated mRNAs, 14,247 annotated lncRNAs and 1290 novel lncRNAs were identified. A total of 227 annotated lncRNAs and 13 novel lncRNAs were significantly and differentially expressed after viral infection. The Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) databases were used to identify genes adjacent to the lncRNAs, and it was found that these lncRNAs were enriched in pathways related to immune and antiviral responses. Next, we selected candidate lncRNAs and their predicted target genes for study. RT-qPCR demonstrated that these lncRNAs and genes were differentially expressed after PEDV infection. Our study investigated the function of lncRNAs involved in PEDV infection, providing new insight into the pathogenic mechanisms of PEDV.




Similar content being viewed by others
Availability of data and materials
The genetic data presented in this paper are publicly available in the GenBank database under accession no. MG983755.
Abbreviations
- PED:
-
Porcine epidemic diarrhea
- PEDV:
-
Porcine epidemic diarrhea virus
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- RT-qPCR:
-
Real-time quantitative PCR
- RNA-Seq:
-
RNA sequencing
- CPC:
-
Coding Potential Calculator
- MOI:
-
Multiplicity of infection
- PLA2G4C:
-
Cytosolic phospholipase A2 gamma
- TIRAP:
-
Toll/interleukin 1 receptor domain-containing adaptor protein
- CLIP170:
-
Cytoplasmic linker protein 170
- IL6R:
-
Interleukin-6 receptor
- IL6:
-
Interleukin-6
- STAT3:
-
Signal transducer and activator of transcription 3
References
Wood EN (1977) An apparently new syndrome of porcine epidemic diarrhoea. Vet Rec 100:243–244
Pensaert MB, de Bouck P (1978) A new coronavirus-like particle associated with diarrhea in swine. Arch Virol 58:243–247
Sun D, Wang X, Wei S, Chen J, Feng L (2016) Epidemiology and vaccine of porcine epidemic diarrhea virus in China: a mini-review. J Vet Med Sci 78:355–363
Song D, Park B (2012) Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes 44:167–175
Fan B, Jiao D, Zhao X, Pang F, Xiao Q, Yu Z et al (2017) Characterization of Chinese porcine epidemic diarrhea virus with novel insertions and deletions in genome. Sci Rep 7:44209
Lee S, Lee C (2017) Complete genome sequence of a novel S-insertion variant of porcine epidemic diarrhea virus from South Korea. Arch Virol 162:2919–2922
Suzuki T, Shibahara T, Yamaguchi R, Nakade K, Yamamoto T, Miyazaki A et al (2016) Pig epidemic diarrhoea virus S gene variant with a large deletion non-lethal to colostrum-deprived newborn piglets. J Gen Virol 97:1823–1828
Costa FF (2010) Non-coding RNAs: meet thy masters. BioEssays 32:599–608
Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Natrev Genet 10:155–159
Imam H, Bano AS, Patel P, Holla P, Jameel S (2015) The lncRNA NRON modulates HIV-1 replication in a NFAT-dependent manner and is differentially regulated by early and late viral proteins. Sci Rep 5:8639
Xiong Y, Yuan J, Zhang C, Zhu Y, Kuang X, Lan L et al (2015) The STAT3-regulated long non-coding RNA Lethe promote the HCV replication. Biomed Pharmacother 72:165–171
Ouyang J, Zhu X, Chen Y, Wei H, Chen Q, Chi X et al (2014) NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe 16:616–626
Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K et al (2014) Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell 53:393–406
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Winterling C, Koch M, Koeppel M, Garcia-Alcalde F, Karlas A, Meyer TF (2014) Evidence for a crucial role of a host non-coding RNA in influenza A virus replication. RNA Biol 11:66–75
Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang Y, Tang GN et al (2013) Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology 57:1882–1892
Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y et al (2012) Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem 287:26302–26311
Zhang Q, Chen CY, Yedavalli VS, Jeang KT (2013) NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. Mbio 4:e512–e596
Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE 7:e30619
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ et al (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515
Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L et al (2007) CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35:W345–W349
Kang YJ, Yang DC, Kong L, Hou M, Meng YQ, Wei L et al (2017) CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res 45:W12–W16
Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL et al (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285
Carrieri C, Laura C, Marta B, Anne B, Silvia Z, Stefania F et al (2012) Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491:454–457
Ding Z, Fang L, Jing H, Zeng S, Wang D, Liu L et al (2014) Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J Virol 88:8936–8945
Zhang Q, Ma J, Yoo D (2017) Inhibition of NF-kappaB activity by the porcine epidemic diarrhea virus nonstructural protein 1 for innate immune evasion. Virology 510:111–126
Zhang Q, Shi K, Yoo D (2016) Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology 489:252–268
Yu L, Dong J, Wang Y, Zhang P, Liu Y, Zhang L et al (2019) Porcine epidemic diarrhea virus nsp4 induces pro-inflammatory cytokine and chemokine expression inhibiting viral replication in vitro. Arch Virol 164:1147–1157
Qian X, Xu C, Zhao P, Qi Z (2016) Long non-coding RNA GAS5 inhibited hepatitis C virus replication by binding viral NS3 protein. Virology 492:155–165
Li J, Chen C, Ma X, Geng G, Liu B, Zhang Y et al (2016) Long noncoding RNA NRON contributes to HIV-1 latency by specifically inducing tat protein degradation. Nat Commun 7:11730
Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A et al (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25:1915–1927
Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G et al (2011) lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477:295–300
He Y, Ding Y, Zhan F, Zhang H, Han B, Hu G et al (2015) The conservation and signatures of lincRNAs in Marek's disease of chicken. Sci Rep 5:15184
Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ et al (2012) Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 22:577–591
Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F et al (2016) A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351:271–275
Stewart A, Ghosh M, Spencer DM, Leslie CC (2002) Enzymatic properties of human cytosolic phospholipase A(2)gamma. J Biol Chem 277:29526–29536
Underwood KW, Song C, Kriz RW, Chang XJ, Knopf JL, Lin LL (1998) A novel calcium-independent phospholipase A2, cPLA2-gamma, that is prenylated and contains homology to cPLA2. J Biol Chem 273:21926–21932
Xu S, Pei R, Guo M, Han Q, Lai J, Wang Y et al (2012) Cytosolic phospholipase A2 gamma is involved in hepatitis C virus replication and assembly. J Virol 86:13025–13037
Narayanan KB, Park HH (2015) Toll/interleukin-1 receptor (TIR) domain-mediated cellular signaling pathways. Apoptosis 20:196–209
Jakka P, Bhargavi B, Namani S, Murugan S, Splitter G, Radhakrishnan G (2018) Cytoplasmic linker protein CLIP170 negatively regulates TLR4 signaling by targeting the TLR adaptor protein TIRAP. J Immunol 200:704–714
Avendano-Tamayo E, Rua A, Parra-Marin MV, Rojas W, Campo O, Chacon-Duque J et al (2019) Evaluation of variants in IL6R, TLR3, and DC-SIGN genes associated with dengue in sampled Colombian population. Biomedical 39:88–101
Revez JA, Bain LM, Watson RM, Towers M, Collins T, Killian KJ et al (2019) Effects of interleukin-6 receptor blockade on allergen-induced airway responses in mild asthmatics. Clin Transl Immunol 8:e1044
Yoshida K, Chambers I, Nichols J, Smith A, Saito M, Yasukawa K et al (1994) Maintenance of the pluripotential phenotype of embryonic stem cells through direct activation of gp130 signalling pathways. Mech Dev 45:163–171
Nichols J, Chambers I, Smith A (1994) Derivation of germline competent embryonic stem cells with a combination of interleukin-6 and soluble interleukin-6 receptor. Exp Cell Res 215:237–239
Shen XH, Cui XS, Lee SH, Kim NH (2012) Interleukin-6 enhances porcine parthenote development in vitro, through the IL-6/Stat3 signaling pathway. J Reprod Dev 58:453–460
Song Y, Yang X, Shen Y, Wang Y, Xia X, Zhang AM (2019) STAT3 signaling pathway plays importantly genetic and functional roles in HCV infection. Mol Genet Genom Med 7(8):e821. https://doi.org/10.1002/mgg3.821
Funding
This work was supported by the National Key Research and Development Program of China (2018YFD0501102), Guangdong Rural Revitalization Strategy Program (201817SY0002), National Key Technologies R&D Program (2015BAD12B02-5), Henan Scale Pig Farm Major Disease Purification and Innovative Technology Team, the Henan Science and Technology Project (182102110037), and Key and Cultivation Discipline of Xinyang Agriculture and Forestry University (ZDXK201702).
Author information
Authors and Affiliations
Contributions
LY, JD, and YL contributed equally to the work; LY and YL performed the assays and data analysis; LZ, PL, and LW performed the sequence alignment; LH contributed to the experimental design and revision of manuscript; and JD and CS wrote the manuscript. All authors reviewed and approved the final form of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethics approval and consent to participate
All of the samples were collected according to the animal ethics regulations of the National Engineering Center for Swine Breeding Industry (NECSBI 2015-16).
Additional information
Handling Editor: Sheela Ramamoorthy.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yu, L., Dong, J., Liu, Y. et al. Genome-wide analysis of long noncoding RNA profiles in Vero cells infected with porcine epidemic diarrhea virus. Arch Virol 165, 1969–1977 (2020). https://doi.org/10.1007/s00705-020-04694-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04694-4