Abstract
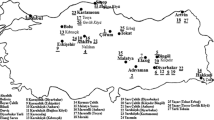
Crop wild relatives of rice (Oryza sativa L.) are an important reservoir of genes for broadening the genetic base of commercial varieties. India is the centre of genetic diversity of some of the wild as-well-as the cultivated rice species. In order to collect genetic diversity, explorations were undertaken in six states located in different agro-ecological zones of India. Forty-four accessions belonging to four wild species (Oryza rufipogon Griff., Oryza nivara Sharma et Shastry, Oryza officinalis Wall. ex Watt and Oryza sativa L. f. spontanea Roshev.) including two accessions of cultivated rice (Oryza sativa L. subsp. indica Kato) were collected from different ecological niches. The rice accessions were characterized using 28 morphological descriptors, which included 15 qualitative and 13 quantitative traits. Wild rice accessions displayed a broad range of phenotypic variation for plant height, main heading days, days to 50% flowering, flag-leaf length and width, panicle shape and size, peduncle length, awning pattern, awn length and colour, seed length and width, seed length/width ratio and 100-seed weight. Cluster analysis based on Jaccard’s similarity coefficients derived from 28 morphological traits grouped 44 accessions into three clades. DNA sequencing revealed that internal transcribed spacer (ITS) region of 18S–5.8S–28S nuclear ribosomal cistron varied from 574 to 577 bp with mean of 576.1 bp in AA-genome species. GC content was high in ITS region, which varied from 69.57 to 70.36% with mean value of 69.99%. Neighbour-joining phylogenetic tree distinguished Oryza sativa–nivara–rufipogon complex from other species of AA-genome. The wild germplasm characterized in this study could be utilized in the genetic enhancement of cultivated rice.






Similar content being viewed by others
References
Aggarwal RK, Brar DS, Khush GS (1997) Two new genomes in the Oryza complex identified on the basis of molecular divergence analysis using total genomic DNA hybridization. Mol Gen Genet 254:1–12
Aggarwal RK, Brar DS, Nandi S, Huang N, Khush GS (1999) Phylogenetic relationships among Oryza species revealed by AFLP markers. Theor Appl Genet 98:1320–1328
Almarazi T, Roux C, Maumont S, Durrieu G (2002) Phylogenetic relationships among smut fungi parasitizing dicotyledons based on ITS sequence analysis. Mycol Res 106:541–548. https://doi.org/10.1017/S0953756202006019
Alvarez I, Wendel JF (2003) Ribosomal ITS sequences and plant phylogenetic inference. Mol Phylogenet Evol 29:417–434. https://doi.org/10.1016/s1055-7903(03)00208-2
Ammiraju JSS, Fan C, Yu Y, Song X, Cranston KA, Pontaroli AC, Lu F, Sanyal A, Jiang N, Rambo T, Currie J, Collura K, Talag J, Bennetzen JL, Chen M, Jackson S, Wing RA (2010) Spatio-temporal patterns of genome evolution in allotetraploid species of the genus Oryza. Plant J 63:430–442. https://doi.org/10.1111/j.1365-313X.2010.04251.x
Arrieta-Espinoza G, Sanchez E, Vargas S, Lobo J, Quesada T, Espinoza AM (2005) The weedy rice complex in Costa Rica. I. Morphological study of relationships between commercial rice varieties, wild Oryza relatives and weedy types. Genet Resour Crop Evol 52:575–587
Bengtsson-Palme J, Ryberg M, Hartmann M, Branco S, Wang Z, Godhe A, DeWit P, Sanchez-Garc M, Ebersberger I, de Sousa F, Amend AS, Jumpponen A, Unterseher M, Kristiansson E, Abarenkov K, Bertrand YJK, Kemal S, Sanli K, Eriksson KM, Vik U, Veldre V, Nilsson RH (2013) Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol 4:914–919. https://doi.org/10.1111/2041-210x.12073
Brar DS, Khush GS (1997) Alien gene introgression in rice. Plant Mol Biol 35:35–47
Brar DS, Khush GS (2003) Utilization of wild species of genus Oryza in rice improvement. In: Nanda JS, Sharma SD (eds) Monograph on genus Oryza. Oxford & IBH Publishing, New Delhi, pp 283–309
Chitrakon S (1995) Characterization, evaluation and utilization of wild rice germplasm in Thailand. Pathum Thani Rice Research Center, Thanya Buri, p 143
Cordesse F, Second G, Delseny M (1990) Ribosomal gene spacer length variability in cultivated and wild rice species. Theor Appl Genet 79:81–88
Dong YB, Pei XW, Yuan QH, Wu HJ, Wang XJ, Jia SR, Peng YF (2010) Ecological, morphological and genetic diversity in Oryza rufipogon Griff. (Poaceae) from Hainan Island, China. Genet Resour Crop Evol 57:915–926
Dover G (1982) Molecular drive: a cohesive mode of species evolution. Nature 299:111–117
Duan S, Lu B, Li Z, Tong J, Kong J, Yao W, Li S, Zhu Y (2007) Phylogenetic analysis of AA-genome Oryza species (Poaceae) based on chloroplast, mitochondrial, and nuclear DNA sequences. Biochem Genet 45:113–129
Feliner GN, Rosselló JA (2007) Review—Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Mol Phylogenet Evol 44:911–919. https://doi.org/10.1016/j.ympev.2007.01.013
Fenton B, Malloch G, Germa F (1998) A study of variation in rDNA ITS regions shows that two haplotypes coexist within a single aphid genome. Genome 41:337–345
Gaikwad KB, Singh N, Bhatia D, Kaur R, Bains NS, Bharaj TS, Singh K (2014) Yield-enhancing heterotic QTL transferred from wild species to cultivated rice Oryza sativa L.. PLoS One 9:1–11. https://doi.org/10.1371/journal.ponr.0096939
Ge S, Sang T, Lu BR, Hong DY (1999) Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proc Natl Acad Sci USA 96:14400–14405
Honsy M, Hijri M, Passerieux E, Dulieu H (1999) rDNA units are highly polymorphic in Scutellospora castanea (Glomales, Zygomycetes). Gene 226:61–71
Hore DK, Sharma BD (1993) Wild rice genetic resources of North-East India. Indian J Plant Genet Resour 6:27–32
Joseph L, Kuriachan P, Kalyanaraman K (1999) Collection and evaluation of the tetraploid Oryza officinalis Wall ex Watt (O. malampuzhaensis Krish.et Chand.) endemic to Western Ghats, India. Genet Resour Crop Evol 46:531–541
Juliano AB, Naredo MEB, Jackson MT (1998) Taxonomic status of Oryza glumaepatula Steud. I. Comparative morphological studies of New World diploids and Asian AA genomes species. Genet Resour Crop Evol 45:197–203
Khush GS (1977) Disease and insect resistance in rice. Adv Agron 29:265–341
Khush GS (1989) Multiple disease and insect resistance for increased yield stability in rice. Progress in irrigated rice research. International Rice Research Institute, Los Baños, pp 79–92
Khush GS (1997) Origin, dispersal, cultivation and variation of rice. Plant Mol Biol 35:25–34
Khush GS (2005) What it will take to feed 5.0 billion rice consumers in 2030? Plant Mol Biol 59:1–6
Khush GS (2013) Strategies for increasing the yield potential of cereals: case of rice as an example. Plant Breed 132:433–436
Khush GS, Virk PS (2000) Rice breeding: achievements and future strategies. Crop Improv 27:115–144
Li QQ, Zhou SD, Huang DQ, He XJ, Wei XQ (2016) Molecular phylogeny, divergence time estimates and historical biogeography within one of the world’s largest Monocot genera. AoB Plants. https://doi.org/10.1093/aobpla/plw041
Liang F, Deng Q, Wang Y, Xiong Y, Jin D, Li J, Wang B (2004) Molecular marker-assisted selection for yield-enhancing genes in the progeny of “9311 × O. rufipogon” using SSR. Euphytica 139:159–165
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Lu BR, Sharma SD (2003) Exploration, collection and conservation of wild Oryza species. In: Nanda JS, Sharma SD (eds) Monograph on genus Oryza. Oxford & IBH Publishing, New Delhi, pp 263–282
Lu F, Ammiraju JSS, Sanyal A, Zhang S, Song R, Chen J, Li G, Sui Y, Song X, Cheng Z, de Oliveira AC, Bennetzen JL, Jackson SA, Wing RA, Chen M (2009) Comparative sequence analysis of Monoculm1-orthologous regions in 14 Oryza genomes. Proc Natl Acad Sci USA 106:2071–2076
McCouch SR, McNally KL, Wang W, Hamilton RS (2012) Genomics of gene banks: a case study in rice. Am J Bot 99:407–423
McCouch SR, Baute GJ, Bramel P, Bretting PK, Buckler E et al (2013) Feeding the future. Nature 499:23–24
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8:4321–4325
Naumann A, Navarro-González M, Sánchez-Hernández O, Hoegger PJ, Kües U (2007) Correct identification of wood-inhabiting fungi by ITS analysis. Curr Trends Biotechnol Pharm 1:41–61
Ohtsubo H, Cheng C, Ohsawa I, Tsuchimoto S, Ohtsubo E (2004) Rice retroposon ρ-SINE1 and origin of cultivated rice. Breed Sci 54:1–11. https://doi.org/10.1270/jsbbs.54.1
Patra BC, Dhua SR, Marndi BC, Nayak PK, Swain P, Kumar GAK, Singh K (2008) Exploration, collection, characterization, evaluation and conservation of wild rice germplasm of east India. Oryza 45:98–102
Rohlf FJ (1998) NTSYSpc: numerical taxonomy and multivariate analysis system version 2.02h. Exeter Software, New York
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Samal R, Roy PS, Sahoo A, Kar MK, Patra BC, Marndi BC, Gundimeda JNR (2018) Morphological and molecular dissection of wild rices from eastern India suggests distinct speciation between O. rufipogon and O. nivara populations. Sci Rep 8:2773. https://doi.org/10.1038/s41598-018-20693-7
Sanchez PA, Wing RA, Brar DS (2014) The wild relatives of rice: genomes and genomics. In: Zhang Q, Wing RA (eds) Genetics and genomics of rice, plant genetics and genomics: crops and models, vol 5. Springer Science-Business Media, New York, pp 9–25. https://doi.org/10.1007/978-1-4614-7903-1-2
Sarla N, Bobba S, Siddiq EA (2003) ISSR and SSR markers based on AG and GA repeats delineate geographically diverse Oryza nivara accessions and reveal rare alleles. Curr Sci 84:683–690
Shen XH, Yan S, Huang RL, Zhu S, Xiong HL, Shen LJ (2013) Development of novel cytoplasmic male sterile source from Dongxiang wild rice (Oryza rufipogon). Rice Sci 20:379–382
Singh A, Singh B, Panda K, Rai VP, Singh AK, Singh SP, Chouhan SK, Rai V, Singh PK, Singh NK (2013) Wild rices of eastern Indo-Gangetic plains of India constitute two sub-populations harbouring rich genetic diversity. Plant Omics J 6:121–127
Singh BP, Singh B, Mishra S, Kumar V, Singh NK (2016) Genetic diversity and population structure in Indian wild rice accessions. Aust J Crop Sci 10:144–151
Singh B, Singh N, Mishra S, Tripathi K, Singh BP, Rai V, Singh AK, Singh NK (2018) Morphological and molecular data reveal three distinct populations of Indian wild rice Oryza rufipogon Griff. species complex. Front Plant Sci 9:1–18. https://doi.org/10.3389/fpls.2018.00123
Stein JC, Yu Y, Copetti D, Zwickl DJ, Zhang L, Zhang C et al (2018) Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat Genet 50:285–296. https://doi.org/10.1038/s41588-018-0040-0
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 24:4876–4882
Tiwari S, Yadav MC, Dikshit N, Pani DR (2015) Universal rice primer based molecular characterization reveals high level of genetic variation in crop-wild relatives of rice. New Agric 26:319–328
Vaughan DA (1989) The genus Oryza L. Current status of taxonomy. IRRI Res Pap Ser 138:1–21
Vaughan DA (1994) The wild relatives of rice: a genetic resources handbook. International Rice Research Institute, Manila
Vaughan DA, Morishima H (2003) Biosystematics of the genus Oryza. In: Smith CW, Dilday RH (eds) Rice-origin, history, technology and production. Wiley, New York, pp 27–65
Vaughan DA, Morishima H, Kadowaki K (2003) Diversity in the Oryza genus. Curr Opin Plant Biol 6:139–146
Vaughan DA, Kadowaki K-L, Kaga A, Tomooka N (2005) On the phylogeny and biography of the genus Oryza. Breed Sci 55:113–122. https://doi.org/10.1270/jsbbs.55.113
Veasey EA, Silva EFD, Schammass EA, Oliveira GCX, Ando A (2008) Morpho-agronomic genetic diversity in American wild rice species. Braz Arch Biol Technol Int J 51:95–104
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press Inc, San Diego, pp 315–322
Xiao J, Grandillo S, Ahn SN, McCouch SR, Tanksley SD, Li J, Yuan L (1996) Genes from wild rice improve yield. Nature 384:223–224
Xiao J, Li J, Grandillo S, Ahn SN, Yuan L, Tanksley SD, McCouch SR (1998) Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Oryza rufipogon. Genetics 150:899–909
Xu X, Liu X, Ge S, Jensen JD, Hu F, Li X et al (2012) Resequencing of 50 accessions of cultivated and wild rice yields markers for identifying agronomically important traits. Nat Biotechnol 30:105–111. https://doi.org/10.1038/nbt.2050
Yan S, Zhu S, Mao LH, Huang RL, Xiong HL, Shen LJ, Shen XH (2017) Molecular identification of the cytoplasmic male sterile source from Dongxiang wild rice (Oryza rufipogon Griff.). J Integr Agric 16:1669–1675
Yap IP, Nelson RJ (1996) WINBOOT: a program for performing bootstrap analysis of binary data to determine the confidence limits of UPGMA-based dendrograms. In: IRRI discussion Paper Series 14, Manila, Philippines
Zamora A, Barboza C, Lobo J, Espinoza AM (2003) Diversity of native rice (Oryza Poaceae) species of Costa Rica. Genet Resour Crop Evol 50:855–870
Zhou Y, Zou YP, Hong DY, Zhou JM, Chen SY (1996) ITS1 sequences of nuclear ribosomal DNA in wild rices and cultivated rices of China and their phylogenetic implications. Acta Bot Sin 38:785–791
Acknowledgements
This work was partially funded from Indian Council of Agricultural Research (ICAR) Network project on National Innovations in Climate Resilient Agriculture (NICRA) entitled “Focused collection of germplasm from climatic hotspots in wheat and rice, and characterization and evaluation for heat and drought tolerance” with scheme and project codes 13921 and 1006607, respectively. The authors are thankful to Director, ICAR-National Bureau of Plant Genetic Resources, New Delhi for providing research facilities for carrying out the partial research under institute project and D. P. Semwal for geo-referencing of wild rice accessions on India map.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tiwari, S., Yadav, M.C., Dikshit, N. et al. Morphological characterization and genetic identity of crop wild relatives of rice (Oryza sativa L.) collected from different ecological niches of India. Genet Resour Crop Evol 67, 2037–2055 (2020). https://doi.org/10.1007/s10722-020-00958-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-020-00958-9