Abstract
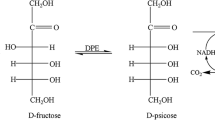
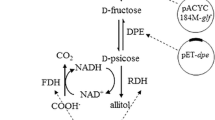
Allitol is a kind of rare sugar alcohol with potential application value. An engineered strain, which simultaneously expressed d-psicose-3-epimerase (DPE), ribitol dehydrogenase (RDH), and formate dehydrogenase (FDH) three enzymes, was constructed by cloning above three genes into one plasmid and transformed into the host E. coli strain, and used as the whole-cell catalysts for biotransformation of allitol from the low-cost substrate of d-fructose. The whole cell allitol biotransformation conditions were optimized. The medium, recombinant gene induction conditions, and the substrate feeding rate for cultivation of the catalytic cells were optimized. Then, the fed-batch culture was made and scaled up to 10 L fermentor. Finally, 63.44 g/L allitol was obtained from 100 g/L d-fructose after 3 h of biotransformation, and the allitol crystals of 99.9% purity were obtained by using cooling recrystallization. The allitol production method developed in this research has high product purity, and is highly efficient, easily scaled up, and suitable for large-scale production of highly purified allitol.









Similar content being viewed by others
References
Takeshita, K., Ishida, Y., Takata, G., & Izumori, K. (2000). Direct production of allitol from D-fructose by a coupling reaction using D-tagatose-3-epimerase, ribitol dehydrogenase and formate dehydrogenase. Journal of Bioscience and Bioengineering, 90(5), 545–548. https://doi.org/10.1016/S1389-1723(01)80038-4.
Oosaka, K. (2009). Possible as monosaccharide laxative of rare sugar alcohols. Yakugaku Zasshi, 129(5), 575–580.
Zhu, Y., Li, H., Liu, P., Yang, J., Zhang, X., & Sun, Y. (2015). Construction of allitol synthesis pathway by multi-enzyme coexpression in Escherichia coli and its application in allitol production. Journal of Industrial Microbiology and Biotechnology, 42(5), 661–669. https://doi.org/10.1007/s10295-014-1578-1.
Hassanin, H. A. M., Mu, W., Koko, M. Y. F., Zhang, T., Masamba, K., & Jiang, B. (2017). Allitol: production, properties and applications. International Journal of Food Science & Technology, 52(1), 91–97. https://doi.org/10.1111/ijfs.13290.
Izumori, K. (2002). Bioproduction strategies for rare hexose sugars. Naturwissenschaften, 89(3), 120–124. https://doi.org/10.1007/s00114-002-0297-z.
Granström, T. B., Takata, G., Tokuda, M., & Izumori, K. (2004). Izumoring: a novel and complete strategy for bioproduction of rare sugars. Journal of Bioscience and Bioengineering, 97(2), 89–94. https://doi.org/10.1016/s1389-1723(04)70173-5.
Izumori, K. (2006). Izumoring: a strategy for bioproduction of all hexoses. Journal of Biotechnology, 124(4), 717–722. https://doi.org/10.1016/j.jbiotec.2006.04.016.
Wu, S. H., Luo, X. D., Ma, Y. B., Liu, J. K., Wu, D. G., Zhao, B., Lu, Y., & Zheng, Q. T. (2000). Two novel secoergosterols from the fungus Tylopilus plumbeoviolaceus. Journal of Natural Products, 63(4), 534–536. https://doi.org/10.1021/np990494h.
Zeng, Y., Dou, D., Zhang, Y., Zhang, L., & Sun, Y. (2014). Rare sugars and antioxidants in Itea virginica, Itea oblonga Hand.-Mazz.,and Itea yunnanensis Franch Leaves. International Journal of Food Properties, 18(11), 2549–2560. https://doi.org/10.1080/10942912.2014.917099.
Jumde, V. R., Eisink, N. N., Witte, M. D., & Minnaard, A. J. (2016). C3 epimerization of glucose, via regioselective oxidation and reduction. The Journal of Organic Chemistry, 81(22), 11439–11443. https://doi.org/10.1021/acs.joc.6b02074.
Kim, H. J., Hyun, E. K., Kim, Y. S., Lee, Y. J., & Oh, D. K. (2006). Characterization of an Agrobacterium tumefaciens d-psicose 3-epimerase that converts d-fructose to d-psicose. Applied and Environmental Microbiology, 72(2), 981–985. https://doi.org/10.1128/AEM.72.2.981-985.2006.
Hassanin, H. A., Wang, X., Mu, W., Zhang, T., & Jiang, B. (2016). Cloning and characterization of a new ribitol dehydrogenase from Providencia alcalifaciens RIMD 1656011. Journal of the Science of Food and Agriculture, 96(8), 2917–2924. https://doi.org/10.1002/jsfa.7589.
Hassanin, H. A. M., Letsididi, R., Koko, M. Y. F., Mu, W., Elferga, A., & Jiang, B. (2016). Synthesis of allitol from D-psicose using ribitol dehydrogenase and formate dehydrogenase. Tropical Journal of Pharmaceutical Research, 15(12), 2701–2708. https://doi.org/10.4314/tjpr.v15i12.23.
He, X., Zhou, X., Yang, Z., Xu, L., Yu, Y., Jia, L., & Li, G. (2015). Cloning, expression and purification of d-tagatose 3-epimerase gene from Escherichia coli JM109. Protein Expression and Purification, 114, 77–81. https://doi.org/10.1016/j.pep.2015.06.015.
Muniruzzaman, S., Tokunaga, H., & Izumori, K. (1995). Conversion of d-psicose to allitol by Enterobacter agglomerans strain 221e. Journal of Fermentation and Bioengineering, 79(4), 323–327. https://doi.org/10.1016/0922-338X(95)93989-W.
Han, W., Zhu, Y., Men, Y., Yang, J., Liu, C., & Sun, Y. (2014). Production of allitol from D-psicose by a novel isolated strain of Klebsiella oxytoca G4A4. Journal of Basic Microbiology, 54(10), 1073–1079. https://doi.org/10.1002/jobm.201300647.
Hassanin, H. A., Eassa, M. A., & Jiang, B. (2018). Facile synthesis of bioactive allitol from D-psicose by coexpression of ribitol dehydrogenase and formate dehydrogenase in Escherichia coli. Journal of Food Bioactives, 4, 117–122. https://doi.org/10.31665/JFB.20xx.000xx.
Lu, F., Xu, W., Zhang, W., Guang, C., & Mu, W. (2019). Polyol dehydrogenases: intermediate role in the bioconversion of rare sugars and alcohols. Applied Microbiology and Biotechnology, 103(16), 6473–6481. https://doi.org/10.1007/s00253-019-09980-z.
Takeshita, K., Suga, A., Takada, G., & Izumori, K. (2000). Mass production of D-psicose from D-fructose by a continuous bioreactor system using immobilized D-tagatose 3-epimerase. Journal of Bioscience and Bioengineering, 90(4), 453–455. https://doi.org/10.1016/s1389-1723(01)80018-9.
Choi, J. G., Ju, Y. H., Yeom, S. J., & Oh, D. K. (2011). Improvement in the thermostability of D-psicose 3-epimerase from Agrobacterium tumefaciens by random and site-directed mutagenesis. Applied and Environmental Microbiology, 77(20), 7316–7320. https://doi.org/10.1128/AEM.05566-11.
Li, C., Lin, J., Guo, Q., Zhang, C., Du, K., Lin, H., & Lin, J. (2018). D-psicose 3-epimerase secretory overexpression, immobilization, and d-psicose biotransformation, separation and crystallization. Journal of Chemical Technology and Biotechnology, 93(2), 350–357. https://doi.org/10.1002/jctb.5360.
Li, C., Zhang, C., Lin, J., Gao, L., Lin, H., & Lin, J. (2018). Enzymatic fructose removal from D-psicose bioproduction model solution and the system modeling and simulation. Journal of Chemical Technology and Biotechnology, 93(5), 1249–1260. https://doi.org/10.1002/jctb.5483.
Makower, B., & Dye, W. B. (1956). Sugar crystallization, equilibrium moisture content and crystallization of amorphous sucrose and glucose. Journal of Agricultural and Food Chemistry, 4(1), 72–77. https://doi.org/10.1021/jf60059a010.
Restaino, O. F., Bhaskar, U., Paul, P., Li, L., De Rosa, M., Dordick, J. S., & Linhardt, R. J. (2013). High cell density cultivation of a recombinant E. coli strain expressing a key enzyme in bioengineered heparin production. Applied Microbiology and Biotechnology, 97(9), 3893–3900. https://doi.org/10.1007/s00253-012-4682-z.
Faust, G., Janzen, N. H., Bendig, C., Romer, L., Kaufmann, K., & Weuster-Botz, D. (2014). Feeding strategies enhance high cell density cultivation and protein expression in milliliter scale bioreactors. Biotechnology Journal, 9(10), 1293–1303. https://doi.org/10.1002/biot.201400346.
Chen, Y., Li, L., Long, L., & Ding, S. (2018). High cell-density cultivation of phenolic acid decarboxylase-expressing Escherichia coli and 4-vinylguaiacol bioproduction from ferulic acid by whole-cell catalysis. Journal of Chemical Technology and Biotechnology, 93(8), 2415–2421. https://doi.org/10.1002/jctb.5590.
Priebe, X., Daschner, M., Schwab, W., & Weuster-Botz, D. (2018). Rational selection of biphasic reaction systems for geranyl glucoside production by Escherichia coli whole-cell biocatalysts. Enzyme and Microbial Technology, 112, 79–87. https://doi.org/10.1016/j.enzmictec.2017.11.003.
Zhu, Y., Men, Y., Bai, W., Li, X., Zhang, L., Sun, Y., & Ma, Y. (2012). Overexpression of d-psicose 3-epimerase from Ruminococcus sp. in Escherichia coli and its potential application in d-psicose production. Biotechnology Letters, 34(10), 1901–1906. https://doi.org/10.1007/s10529-012-0986-4.
Zhang, W., Fang, D., Xing, Q., Zhou, L., Jiang, B., & Mu, W. (2013). Characterization of a novel metal-dependent D-psicose 3-epimerase from Clostridium scindens 35704. PLoS One, 8(4), e62987. https://doi.org/10.1371/journal.pone.0062987.
Ferenci, T., & Kornberg, H. L. (1973). The utilization of fructose by Escherichia coli. Properties of a mutant defective in fructose-1-phosphate kinase activety. Biochemical Journal, 132(2), 341–347. https://doi.org/10.1042/bj1320341.
Ferenci, T., & Kornberg Hans, L. (1974). The role of phosphotransferase-mediated syntheses of fructose 1-phosphate and fructose 6-phosphate in the growth of Escherichia coli on fructose. Proceedings of the Royal Society B: Biological Sciences, 187(1087), 105–119. https://doi.org/10.1098/rspb.1974.0065.
Kornberg, H. L., Lambourne, L. T. M., & Sproul, A. A. (2000). Facilitated diffusion of fructose via the phosphoenolpyruvate/glucose phosphotransferase system of Escherichia coli. Proceedings of the National Academy of Sciences, 97(4), 1808–1812. https://doi.org/10.1073/pnas.97.4.1808.
Pastor, J. M., Borges, N., Pagan, J. P., Castano-Cerezo, S., Csonka, L. N., Goodner, B. W., Reynolds, K. A., Goncalves, L. G., Argandona, M., Nieto, J. J., Vargas, C., Bernal, V., & Canovas, M. (2019). Fructose metabolism in Chromohalobacter salexigens: interplay between the embden-meyerhof-parnas and entner-doudoroff pathways. Microbial Cell Factories, 18(1), 134–148. https://doi.org/10.1186/s12934-019-1178-x.
Ji, X. J., Huang, H., Du, J., Zhu, J. G., Ren, L. J., Li, S., & Nie, Z. K. (2009). Development of an industrial medium for economical 2,3-butanediol production through co-fermentation of glucose and xylose by Klebsiella oxytoca. Bioresource Technology, 100(21), 5214–5218. https://doi.org/10.1016/j.biortech.2009.05.036.
Ye, Q., Li, X., Yan, M., Cao, H., Xu, L., Zhang, Y., Chen, Y., Xiong, J., Ouyang, P., & Ying, H. (2010). High-level production of heterologous proteins using untreated cane molasses and corn steep liquor in Escherichia coli medium. Applied Microbiology and Biotechnology, 87(2), 517–525. https://doi.org/10.1007/s00253-010-2536-0.
Borji, A., & Jourani, A. (2018). Spectrophotometry as a method for the determination of solubility of sucrose in water and metastable zone width of its aqueous solutions. Crystal Research and Technology, 53(6), 1700123–1700128. https://doi.org/10.1002/crat.201700123.
Shao, X. F., Yang, S., Wang, C., Yang, Y. J., Wang, W. J., Zeng, Y., & Fan, L. W. (2019). Screening of sugar alcohols and their binary eutectic mixtures as phase change materials for low-to-medium temperature thermal energy storage. (II): Isothermal melting and crystallization behaviors. Energy, 180, 572–583. https://doi.org/10.1016/j.energy.2019.05.109.
Singh, K., Gupta, S. P., Kumar, A., & Kumar, A. (2019). The effect of high intensity ultrasound (HIU) on the kinetics of crystallization of sucrose: elimination of latent period. Ultrasonics-Sonochemistry, 52, 19–24. https://doi.org/10.1016/j.ultsonch.2018.05.030.
Kaup, B., Bringer-Meyer, S., & Sahm, H. (2004). Metabolic engineering of Escherichia coli: construction of an efficient biocatalyst for d-mannitol formation in a whole-cell biotransformation. Applied Microbiology and Biotechnology, 64(3), 333–339. https://doi.org/10.1007/s00253-003-1470-9.
Lin, J. Q., Lee, S. M., & Koo, Y. M. (2001). Hydrolysis of paper mill sludge using an improved enzyme system. Journal of Microbiology and Biotechnology, 11(3), 362–368.
Alekseeva, A. A., Fedorchuk, V. V., Zarubina, S. A., Sadykhov, E. G., Matorin, A. D., Savin, S. S., & Tishkov, V. I. (2015). The role of Ala198 in the stability and coenzyme specificity of bacterial formate dehydrogenases. Acta Naturae, 7(1), 60–69.
Li, C., Lin, J. Q., Gao, L., Lin, H. B., & Lin, J. Q. (2018). Modeling and simulation of enzymatic gluconic acid production using immobilized enzyme and CSTR–PFTR circulation reaction system. Biotechnology Letters, 40(4), 649–657. https://doi.org/10.1007/s10529-018-2509-4.
Huang, R., Chen, H., Zhong, C., Kim, J. E., & Zhang, Y. H. (2016). High-throughput screening of coenzyme preference change of thermophilic 6-phosphogluconate dehydrogenase from NADP+ to NAD+. Scientific Reports, 6, 32644–32644. https://doi.org/10.1038/srep32644.
Acknowledgments
The authors would like to thank Chengjia Zhang, Caiyun Sun from the Core Facilities for Life and Environmental Sciences, State Key Lab of Microbial Technology for help and guidance in the experiments.
Funding
This research was funded by the Key R & D Plan of Shandong Province in 2019 (2019GSF107015), and Shandong Province Science and Technology Development Project (2015GSF121016) of China.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
The authors declare that there are no studies conducted with human participants or animals.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wen, X., Lin, H., Ren, Y. et al. Efficient Allitol Bioproduction from d-Fructose Catalyzed by Recombinant E. coli Whole Cells, and the Condition Optimization, Product Purification. Appl Biochem Biotechnol 192, 680–697 (2020). https://doi.org/10.1007/s12010-020-03359-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-020-03359-x