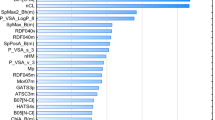
Abstract
Factor Xa (FXa) enzyme has an important role in the blood coagulation system. Disruption in the enzyme function results in the production of blood clots. Therefore, inhibition of the factor Xa with anticoagulant drugs is an important target in thromboembolic therapy. Experimental design of new drugs is a time-consuming and expensive process. Application of molecular modeling is an essential step for designing new and high yield drugs to achieve better results. In this study, 36 aroylguanidine derivatives that inhibit destructive activities by binding to the FXa enzyme were selected. Quantitative Structure-Activity Relationships (QSAR) model was developed for predicting the IC50 parameter for factor Xa inhibitors. QSAR have been constructed by combining Genetic Algorithms with Multiple Linear Regressions (GA-MLR) and Support Vector Machine (SVM). The correlation coefficient (R2) and root mean square error (RMSE) for GA-MLR are 0.895, 0.142 and for SVM are 0.994, 0.032, respectively. The obtained results indicated that the SVM method is superior over the GA-MLR method. Also, molecular docking was used to examine the results more accurately, in which energy interactions were investigated. Molecular docking indicated the binding relationship between the receptor and the ligand. The results showed that reducing the parameters such as electronegativity, and the size of the atoms, as well as increasing the number of loops and groups attached to the structures, is effective in the model.








Similar content being viewed by others
References
Eisenberg PR, Siegel JE, Abendschein DR, Miletich JP (1993) Importance of factor Xa in determining the procoagulant activity of whole-blood clots. J Clin Investig 91:1877–1883
Shi Y, Sitkoff D, Zhang J, Han W, Hu Z, Stein PD, Wang Y, Kennedy LJ, Oconner SP, Ahmad S, Liu EC, Seiler SM, Lam PY, Robl JA, Macor JE, Atwal KS, Zahler R (2007) Amino (methyl) pyrrolidines as novel scaffolds for factor Xa inhibitors. Bioorg Med Chem 17:5952–5958
Xing J, Yang L, Li H, Li Q, Zhao L, Wang X, Zhang Y, Zhou M, Zhou H, Zhang H (2015) Identification of anthranilamide derivatives as potential factor Xa inhibitors: drug design, synthesis and biological evaluation. Eur J Med Chem 95:388–399
Iwatsuki Y, Sato T, Moritani Y, Shigenaga T, Suzuki M, Kawasaki T, Funatsu T, Kaku S (2011) Biochemical and pharmacological profile of darexaban, an oral direct factor Xa inhibitor. Eur J Pharmacol 673:49–55
Verma RP, Hansch C (2010) QSAR modeling of taxane analogues against colon cancer. Eur J Med Chem 45:1470–1477
Choudhari PB, Bhatia MS, Kumbhar SS (2012) 3D QSAR and pharmacophore modeling of selected factor Xa inhibitors. Med Chem Res 21:1427–1432
Jain SV, Ghate M, Bhadoriya KS, Bari SB, Chaudhari A, Borse JS (2012) 2D, 3D-QSAR and docking studies of 1,2,3-thiadiazole thioacetanilides analogues as potent HIV-1 non-nucleoside reverse transcriptase inhibitors. Org Med Chem Lett 2:2–22
Jagiello K, Makurat S, Perec S, Rak J, Puzyn T (2018) Molecular features of thymidine analogues governing the activity of human thymidine kinase. Struct Chem 29:1367–1374
Jagiello K, Sosnowska A, Kar S, Demkowicz S, Dasko M, Leszczynski J, Rachon J, Puzyn T (2017) Geometry optimization of steroid sulfatase inhibitors—the influence on the free binding energy with STS. Struct Chem 28:1017–1032
Fevig JM, Cacciola J, Buriak J, Rossi KA, Knabb RM, Luettgen JM, Wong PC, Bai SA, Wexler RR, Lam PY (2006) Preparation of 1-(4-methoxyphenyl)-1H-pyrazolo [4, 3-d] pyrimidin-7(6H)-ones as potent, selective and bioavailable inhibitors of coagulation factor Xa. Bioorg Med Chem Lett 16:3755–3760
Bhongade BA, Gouripurb VV, Gadada AK (2005) 3D-QSAR CoMFA studies on trypsin-like serine protease inhibitors: a comparative electivity analysis. Bioorg Med Chem 13:2773–2782
Nazare M, Will DW, Matter H (2005) Probing the subpockets of factor Xa reveals two binding modes for inhibitors based on a 2-carboxyindole scaffold: a study combining structure-activity relationship and X-ray crystallography. J Med Chem 48:4511–4525
Dguigui K, Elhallaoui M (2013) 3D-QSAR modeling of substituted thiophene-anthranilamides as potent inhibitors of human factor Xa using quantum chemical descriptors. Int J Sci Res 4:1237–1247
Al-Horani RA, Mehta AY, Desai UR (2012) Potent direct inhibitors of factor Xa based on the tetrahydroisoquinoline scaffold. Eur J Med Chem 54:771e783
Mendel D, Marquart AL, Joseph S (2007) Anthranilamide inhibitors of factor Xa. Bioorg Med Chem Lett 17:4832–4836
Cramer RD, Patterson DE, Bunce JD (1988) Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J Am Chem Soc 110:5959–5967
Vapnik V (1995) The nature of statistical learning theory. Springer-Verlag, New York
Rezaei B, Riahi S, Ebrahimpoor Gorgi A (2020) Molecular investigation of amine performance in the carbon capture process: least squares support vector machine approach. Korean J Chem Eng 37:72–79
Khaheshi Hasnkiadeh SH, Riahi S, Mohammadi-khanaposhti M, Shokrollahzadeh H (2019) Prediction of amines capacity for carbon dioxide absorption based on structural characteristics. Ind Eng Chem Res 58:8763–8771
Mehraein I, Riahi S (2017) The QSPR models to predict the solubility of CO2 in ionic liquids based on least-squares support vector machines and genetic algorithm-multi linear regression. J Mol Liq 225:521–530
Claycamp HG, Sussman NB, Macina O, Rosenkranz HS (1999) Artificial neural networks as statistical tools in SAR/QSAR modeling. AAAI Technical Report SS-99-01, 1-4
Klebe G, Abraham U, Mietzner T (1994) Molecular similarity indices in a comparative analysis (CoMSIA) of drug molecules to correlate and predict their biological activity. J Med Chem 37:4130–4146
Kovalishyn V, Tanchuk V, Charochkina L, Semenuta I, Prokopenko V (2012) Predictive QSAR modeling of phosphodiesterase 4 inhibitors. J Mol Graph Model 32:32–38
Tian Y, Shi Y, Liu X (2012) Recent advances on support vector machines research. Technol Econ Dev Econ 18:5–33
Meng XY, Zhang HX, Mezei M, Cui M (2012) Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des 7:146–157
Shi Y, Li C, Zhang J, Shi M, Huang C, Sitkoff D (2009) Aroylguanidine-based factor Xa inhibitors: the discovery of BMS-344577. Bioorg Med Chem 19:6882–6889
Sepehri B, Rasouli Z, Ghavami R (2016) Molecular docking and QSAR analysis of naphthyridone derivatives as ATAD2 bromodomain inhibitors: application of CoMFA, LS-SVM, and RBF neural network. Med Chem Res 25:2895–2905
Fatemi MH, Heidari A, Gharaghani S (2015) QSAR prediction of HIV-1 protease inhibitory activities using docking derived molecular descriptors. J Theor Biol 369:13–22
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791
Release H (2002) 7.5 for Windows, molecular modeling system, Hypercube. Inc. http://www.Hyper.Com
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JJA, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M (2009) Gaussian 09. Gaussian, Inc., Wallingford
Todeschini R, Consonni V, Mauri A, Pavan M (2002) DRAGON-Software for the calculation of molecular descriptors version 2.1
Todeschini R, Consonni V (2000) Handbook of molecular descriptors. Wiley VCH, Weinheim
Modarresi H, Modarress H, Dearden JS (2007) QSPR model of Henry’s law constant for a diverse set of organic chemicals based on genetic algorithm-radial basis function network approach. Chemosphere 66:2067–2076
Goldberg DE (1989) Genetic algorithms in search, optimization and machine learning. Addison-Wesley Longman, Boston
Yao XJ, Doucet JP, Zhang RS, Chen HF, Liu MC, Hu ZD, Fan BT (2004) Comparative study of QSAR/QSPR correlations using support vector machines, radial basis function neural networks and multiple linear regression. J Chem Inf Model 44:1257–1266
Ghaslani D, Es’haghi Gorji Z, Ebrahimpour Gorji A, Riahi S (2017) Descriptive and predictive models for Henry’s law constant of CO2 liquids: a QSPR study. Chem Eng Res Des 120:15–25
Tropsha A, Gramatica P, Gombar VK (2003) The importance of being earnest: validation is the absolute essential for successful application and interpretation of QSPR models. QSAR Comb Sci 22:69–77
Gramatica P (2007) Principles of QSAR models validation: internal and external. QSAR Comb Sci 26:694–701
Netzeva TI, Worth AP, Aldenberg T, Benigni R, Cronin MT, Gramatica P, Jaworska JS, Kahn S (2005) Current status of methods for defining the applicability domain of (quantitative) structure–activity relationships. The report and recommendations of ECVAM Workshop 52. Altern Lab Anim 33:155–173
Saiz-Urra L, Gonzalez MP, Fall Y, Gomez G (2007) Quantitative structure-activity relationship studies of HIV-1 integrase inhibition. 1. GETAWAY descriptors. Eur J Med Chem 42:64–10
Xinliang Y, Xianwei H (2017) A quantitative relationship between Tgs and chain segment structures of polystryrenes. Plimeros, 27: no. 1 Sao Carlos
Fatemi M, Gharaghani S (2007) A novel QSAR model for prediction of apoptosis-inducing activity of 4-aryl-4-H-chromenes based on support vector machine. Bioorg Med Chem 15:7746–7754
Klein CT, Kaiser D, Ecker G (2004) Topological distance based 3D descriptors for use in QSAR and diversity analysis. J Chem Inf Comput Sci 44:200–209
Hemmer MC, Steinhauer V, Gasteiger J (1999) Deriving the 3D structure of organic molecules from their infrared spectra. Vib Spectrosc 19:151–164
Schuur JH, Selzer P, Gasteiger J (1996) The coding of the three-dimensional structure of molecules by molecular transforms and its application to structure-spectra correlations and studies of biological activity. J Chem Inf Comput Sci 36:334–344
Katritzky AR, Pacureanu LM, Slavov S, Dobchev DA, Karelson M (2006) QSAR study of antiplatelet agents. Bioorg Med Chem 14:7490–7500
Xu C, Ren Y (2015) Molecular modeling studies of [6,6,5] tricyclic fused oxazolidinones as FXa inhibitors using 3D- QSAR, topomer CoMFA, molecular docking and molecular dynamics simulations. Bioorg Med Chem Lett 25:4522–4528
Acknowledgments
The authors gratefully acknowledge the support of the Institute of Petroleum Engineering (IPE), University of Tehran.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ramandi, M., Riahi, S., Rahimi, H. et al. Molecular docking, linear and nonlinear QSAR studies on factor Xa inhibitors. Struct Chem 31, 2023–2040 (2020). https://doi.org/10.1007/s11224-020-01535-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-020-01535-7