Abstract
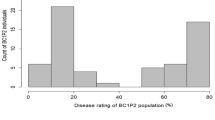
Watermelon [Citrullus lanatus (Thunb.) Matsum & Nakai] is an important cucurbit crop cultivated worldwide. In an effort to map WBNV resistance, the current study consisted of two populations derived from a WBNV resistant accession, IIHR-82 belonging to Citrullus lanatus var. citroides (Syn. Citrullus amarus). The first population is a backcross inbred population (BC1F6) of 141 families (Pop I) that was developed by crossing IIHR-82 to an elite cultivar Arka Manik. A resistant family, BIL-53 belonging to Pop I was crossed to IIHR-140 to develop F3 population, consisting of 112 families (Pop II). Each of these populations was evaluated for WBNV incidence for two years under natural epiphytotic conditions. A significant correlation was observed for various resistance traits across years and populations. The frequency distribution graphs of Area Under Disease Progress Curve for WBNV incidence in both the populations exhibited a continuous distribution, indicating that trait is quantitative in nature. A total of 163 markers were mapped for the Pop I which spread onto 15 linkage groups (LGs) spanning a total map length of 3310.17 cM with a mean marker interval of 20.31 cM. The linkage map for the Pop II was constructed with 135 markers spread over 12 linkage groups spanning a total length of 1965.53 cM with an average interval between markers of 14.56 cM.The QTL analysis for WBNV resistance related traits revealed 14 major QTL's for Pop I and 19 for Pop II with a maximum PVE upto 21.20%. Multi-trait QTL regions were observed on LG 3b for Pop I and LG 2, LG 4, LG 7 and LG 8 for Pop II. Importantly, we observed common QTL regions for plant survival on LG 2 and PDI on LG 3 and LG 8 for both the populations. As this is the first attempt to map QTL's for WBNV resistance, the results obtained in the present study may provide a guide for fine mapping multi-trait QTL regions.






Similar content being viewed by others
References
Adams MJ, Lefkowitz EJ, King AM, Harrach B, Harrison RL, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Mushegian AR, Nibert M (2017) Changes to taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses. Arch Virol 162:2505–2538
Anonymous (2012) Cucurbit Genomics Database. Available at https://www.icugi. org
Branham SE, Wechter WP, Ling KS, Chanda B, Massey L, Zhao G, Guner N, Bello M, Kabelka E, Fei Z, Levi A (2019) QTL mapping of resistance to Fusarium oxysporum f. sp. niveum race 2 and Papaya ringspot virus in Citrullus amarus.Theor Appl Genet. https://doi.org/10.1007/s00122-019-03500-3
Campbell CL, Madden LV (1990) Introduction to plant disease epidemiology. Wiley, New York
Cheng Y, Luan F, Wang X, Gao P, Zhu Z, Liu S, Baloch AW, Zhang Y (2016) Construction of a genetic linkage map of watermelon (Citrullus lanatus) using CAPS and SSR markers and QTL analysis for fruit quality traits. Sci Hort 202:25–31. https://doi.org/10.1016/j.scienta.2016.01.004
Collard BCY, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Phil Trans R Soc B 363:557–572. https://doi.org/10.1098/rstb.2007.2170
Dong W, Wu D, Li G, Wu D, Wang Z (2018) Next-generation sequencing from bulked segregant analysis identifies a dwarfism gene in watermelon. Sci Rep 8:2908. https://doi.org/10.1038/s41598-018-21293-1
Dou J, Zhao S, Lu X, He N, Zhang L, Ali A, Kuang H, Liu W (2018) Genetic mapping reveals a candidate gene (ClFS1) for fruit shape in watermelon (Citrullus lanatus L.). Theor Appl Genet. https://doi.org/10.1007/s00122-018-3050-5
FAO (2016) Food and Agricultural organization (https://www.faostat.fao.org), Rome, Italy. Retrived 10 Febraury 2018
Gimode W, Clevenger J, McGregor C (2020) Fine-mapping of a major quantitative trait locus Qdff3-1 controlling flowering time in watermelon. Mol Breed 40:3
Guo S, Xu Y, Zhang H, Gong G, Huang S, Yi H, Wu M, Zheng Y, Fei Z (2010) Latest advances in watermelon genomics (In Proceedings of the 4th IS on Cucurbits Ed.: Xiaowu Sun). Acta Hort 871:599–606
Guo S, Zhang J, Sun H, Salse J, Lucas WJ, Zhang H, Zheng Y, Mao L, Ren Y, Wang Z, Min J, Guo X, Murat F, Ham B, Zhang Z, Gao S, Huang M, Xu Y, Zhong S, Bombarely A, Mueller LA, Zhao H, He H, Zhang Y, Zhang Z, Huang S, Tan T, Pang E, Lin K, Hu Q, Kuang H, Ni P, Wang B, Liu J, Kou Q, Hou W, Zou X, Jiang J, Gong G, Klee K, Schoof H, Huang Y, Hu X, Dong S, Liang D, Wang J, Wu K, Xia Y, Zhao X, Zheng Z, Xing M, Liang X, Huang B, Lv T, Wang J, Yin Y, Yi H, Li R, Wu M, Levi A, Zhang X, Giovannoni JJ, Wang J, Li Y, Fei Z, Xu Y (2013) The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet 45(1):51–58
Guo S, Zhao S, Sun H, Wang X, Wu S, Lin T, Ren Y, Gao L, Deng Y, Zhang J, Lu X, Zhang H, Shang J, Gong G, Wen C, He N, Tian S, Li M, Liu J, Wang Y, Zhu Y, Jarret R, Levi A, Zhang X, Huang S, Fei Z, Liu W, Xu Y (2019) Resequencing of 414 cultivated and wild watermelon accessions identifies selection for fruit quality traits. Nat Genet 51:1616-1623
Hashizume T, Shimamoto I, Hirai M (2003) Construction of a linkage map and QTL analysis of horticultural traits for watermelon [Citrullus lanatus (Thunb.) Matsum & Nakai] using RAPD, RFLP and ISSR markers. Theor Appl Genet 106:779–785
Hawkins LK, Hawkins TL, Rhodes BB, Jarret RL (2001) Linkage Mapping in a Watermelon Population Segregating for Fusarium Wilt Resistance. J Amer Soc Hort Sci 126(3):344–350
Hobbs HA, Reddy DVR, Rajeshwari R, Reddy AS (1987) Use of direct antigen coating and protein A coating ELISA procedures for detection of three peanut viruses. Plant Dis 71:747–749
Holkar SK, Kumar R, Yogita M, Katiyar A, Jain RK, Mandal B (2017) Diagnostic assays for two closely related tospovirus species, watermelon bud necrosis virus and groundnut bud necrosis virus and identification of new natural hosts. J Plant Biochem Biotechnol 26:43–51
Jahn M, Paran I, Hoffmann K, Radwanski ER, Livingstone KD, Grube RC, Aftergoot E, Lapidot M, Moyer J (2000) Genetic mapping of the Tsw locus for resistance to the Tospovirus tomato spotted wilt virus in Capsicum spp. and Its relationship to the Sw-5 gene for resistance to the same pathogen in tomato. Mol Plant-Microbe Interact 13(6):673–682
Jain RK, Bag S, Umamaheswaran K, Mandal B (2007) Natural infection by tospoviruses of cucurbitaceous and fabaceous vegetable crops in India. J Phytopathol 155:22–25
Jain RK, Mandal B, Pappu HR, Holkar, SK (2015) ICTV Taxonomic Proposal 2014. 005aV. A. v2. Tospovirus_sp. Create 1 New Species in the Genus Tospovirus, Family Bunyaviridae. https://www.ictvonline.org/proposals-14/2014
Jain RK, Pappu HR, Pappu SS, Krishnareddy M, Vani A (1998) Watermelon bud necrosis tospovirusis a distinct virus species belonging to serogroup IV. Arch Virol 143:1637–1644
Joehanes R, Nelson JC (2008) QGene 4.0 An extensible Java QTL-analysis platform. Bioinformatics 24:2788–2789
Keb-Llanes M, Gonzalez G, Chi B, Infante D (2002) A rapid and simple method for small-scale DNA extraction in Agavaceae and other tropical plants. Plant Mol Biol Rep 20:299–303
Khera P, Pandey MK, Wang H, Feng S, Qiao L, Culbreath AK, Kale S, Wang J, Holbrook CC, Zhuang W, Varshney RK, Guo B (2016) Mapping quantitative trait loci of resistance to tomato spotted wilt virus and leaf spots in a recombinant inbred line population of peanut (Arachis hypogaea L.) from SunOleic 97R and NC94022. PLoS ONE 11(7):e0158452. https://doi.org/10.1371/journal.pone.0158452
Kim KH, Hwang JH, Han DY, Park M, Kim S, Choi D, Kim Y, Lee GP, Kim ST, Park YH (2015) Major quantitative trait loci and putative candidate genes for powdery mildew resistance and fruit-related traits revealed by an intraspecific genetic map for watermelon (Citrullus lanatus var. lanatus). PLoS ONE 10(12):e0145665
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugenics 12:172–175
Kumar NKK, Venkatesh N, Kalleshwaraswamy CM, Ranganath HR (2006) Seasonal incidence of thrips and bud necrosis virus on watermelon. Pest Man Hort Ecosyst 12(2):85–92
Kumar R, Mandal B, Geetanjali AS, Jain RK, Jaiwal PK (2010) Genome organisation and sequence comparison suggest intraspecies incongruence in M RNA of Watermelon bud necrosis virus. Arch Virol 155:1361–1365
Kunkalikar SR, Sudarsana P, Arun BM, Rajagopalan P, Chen TC, Yeh SD, Naidu RA, Zehr UB, Ravi KS (2011) Importance and genetic diversity of vegetable-infecting tospoviruses in India. Phytopathology 101:367–376
Lambel S, Lanini B, Vivoda E, Fauve J, Wechter WP, Harris-Shultz KR, Massey L, Levi A (2014) A major QTL associated with Fusarium oxysporum race1 resistance identified in genetic populations derived from closely related watermelon lines using selective genotyping and genotyping-by-sequencing for SNP discovery. Theor Appl Genet 127:2105–2115
Lee HJ, Cho HJ, Lee KA, Lee MS, Shin YS, Harn CH, Yang SG, Nahm SH (2007) Development of sequence-based dna markers for evaluation of phylogenetic relationships in Korean watermelon varieties. J Crop Sci Biotech 10(2):98–105
Levi A, Thies JA, Wechter WP, Harrison HW, Simmons AM, Reddy UK, Nimmakayala P, Fei Z (2013) High frequency oligonucleotides:targeting active gene (HFO-TAG) markers revealed wide genetic diversity among Citrullus spp. accessions useful for enhancing disease or pest resistance in watermelon cultivars. Genet Resour Crop Evol 60:427–440
Levi A, Thomas CE, Keinath AP, Wehner TC (2001a) Genetic diversity among watermelon (Citrullus lanatus and Citrullus colocynthis) accessions. Genet Res Crop Evol 48:559–566
Levi A, Thomas CE, Trebitsh T, Salman A, King J, Karaliaus J, Newman M, Reddy OUK, Xu Y, Zhang X (2006) An extended linkage map for watermelon based on SRAP, AFLP, SSR, ISSR and RAPD markers. J American Soc Hort Sci 131(3):393–402
Levi A, Thomas CE, Zhang X, Joobeur T, Davis A (2002) A genetic linkage map for watermelon derived from a testcross population: (Citrullus lanatus var. citroides × C. lanatus var. lanatus) × Citrullus colocynthis. Theor Appl Genet 105(4):555–563
Levi A, Thomas CE, Zhang X, Joobeur T, Dean RA, Wehner TC, Carle BR (2001b) A genetic linkage map for watermelon based on Randomly Amplified polymorphic DNA (RAPD) markers. J Amer Soc Hort Sci 126:730–737
Levi A, Wechter P, Massey L, Carter L, Hopkins D (2011) An extended genetic linkage map for watermelon based on a testcross and a BC2F2 population. American J Plant Sci 2:93–110
Liu S, Gao P, Wang X, Davis AR, Baloch AM, Luan F (2015) Mapping of quantitative trait loci for lycopene content and fruit traits in Citrullus lanatus. Euphytica 202(3):411–426
MAFW (2017) Horticultural Statistics at a Glance 2017 (https://agricoop.nic.in) Horticulture Statistics Division, Department of Agriculture, Cooperation and Farmers Welfare, Ministry of Agriculture and Farmers Welfare, Government of India
Mandal B, Jain RK, Chaudhary V, Varma A (2003) First report of natural infection of Luffa acutangula by Watermelon bud necrosis virus in India. Plant Dis 87:598
Mandal B, Jain RK, Krishnareddy M, Krishna Kumar NK, Ravi KS, Pappu HR (2012) Emerging problems of Tospoviruses (Bunyaviridae) and their management in the Indian subcontinent. Plant Dis 96(4):468–479
Maule AJ, Caranta C, Boulton MI (2007) Sources of natural resistance to plant viruses: status and prospects. Mol Plant Pathol 8(2):223–231
Nagesh GC, Rao ES, Pitchaimuthu M, Varalakshmi B, Lakshmana Reddy DC, Samuel DK, Rekha A, Krishna Reddy M (2018) Genetic analysis of resistance to watermelon bud necrosis orthotospovirus in watermelon [Citrullus lanatus (Thunb.) Matsum & Nakai]. Plant Breed 137:814–822. https://doi.org/10.1111/pbr.12639
Nimmakayala P, Abburi VL, Bhandary A, Abburi L, Vajja VG, Reddy R, Malkaram S, Venkatramana P, Wijeratne A, Tomason YR, Levi A, Wehner TC, Reddy UK (2014) Use of Vera Code 384-plex assays for watermelon diversity analysis and integrated genetic map of watermelon with single nucleotide polymorphisms and simple sequence repeats. Mol Breed 34:537–548
Park SW, Kim KT, Kang SC, Yang HB (2016) Rapid and practical molecular marker development for rind traits in watermelon. Hort Env Biotech 57:385–391
Prothro J, Sandlin K, Gill R, Bachlava E, White V, Knapp SP, McGregor C (2012) Mapping of the egusi seed trait locus (eg) and quantitative trait loci associated with seed oil percentage in watermelon. J American Soc Hort Sci 137(5):311–315
Rajasekharam T (2010) Biological and molecular characterization and management of Watermelon bud necrosis virus. Ph. D Thesis, UAS, Dharawad
Rebijith KB, Asokan R, Hande HR, Kumar NK (2016) The first report of miRNAs from a thysanopteran insect, Thrips palmi Karny using high-throughput sequencing. PLoS ONE 11:e0163635
Rebijith KB, Asokan R, Krishna Kumar NK, Krishna V, Ramamurthy VV (2012) Development of species-specific markers and molecular differences in mtDNA of Thrips palmi Karny and Scirtothrips dorsalis Hood (Thripidae: Thysanoptera), vectors of tospoviruses Bunyaviridae) in India. Entomol News 122:201–213
Reddy U, Aryal N, Islam-Faridi N, Tomason Y, Levi A, Nimmakayala P (2013) Cytomolecular characterization of rDNA distribution in various Citrullus species using fluorescent in situ hybridization. Genet Resour Crop Evol 60(7):2091–2100. https://doi.org/10.1007/s10722-013-9976-1
Reddy UK, Nimmakayala P, Levi A, Abburi VL, Saminathan T, Tomason YR, Vajja G, Reddy R, Abburi L, Wehner TC, Ronin Y, Karol A (2014) High-resolution genetic map for understanding the effect of genome-wide recombination rate on nucleotide diversity in watermelon. G3 4(11):2219–2230
Ren R, Ray R, Li P, Xu J, Zhang M, Liu G, Yao X, Kilian A, Yang X (2015a) Construction of a high-density DArTseq SNP-based genetic map and identification of genomic regions with segregation distortion in a genetic population derived from a cross between feral and cultivated-type watermelon. Mol Genet Genom 290:1457–1470
Ren Y, Jiao D, Gong G, Zhang H, Guo S, Zhang J, Xu Y (2015b) Genetic analysis and chromosome mapping of resistance to Fusarium oxysporum f. sp. niveum (FON) race 1 and race 2 in watermelon (Citrullus lanatus L.). Mol Breeding 35:183–191
Ren Y, McGregor C, Zhang Y, Gong G, Zhang H, Guo S, Sun H, Cai W, Zhang J, Xu Y (2014) An integrated genetic map based on four mapping populations and quantitative trait loci associated with economically important traits in watermelon (Citrullus lanatus). BMC Plant Biol 14:1471–2229
Ren Y, Zhao H, Kou Q, Jiang J, Guo S, Zhang H, Hou W, Zou X, Sun H, Gong G, Levi A, Xu Y (2012) A High Resolution Genetic Map Anchoring Scaffolds of the Sequenced Watermelon Genome. PLoS ONE 7(1):1–10
Riley DG, Pappu HR (2000) Evaluation of tactics for management of thrips-vectored Tomato spotted wilt virus in tomato. Plant Dis 84(8):847–852
Sandlin K, Prothro J, Heesacker A, Khalilian N, Okashah R, Xiang W, Bachlava E, Caldwell DG, Taylor CA, Seymour DK, White V, Chan E, Tolla G, White C, Safran D, Graham E, Knapp S, McGregor C (2012) Comparative mapping in watermelon [Citrullus lanatus (Thunb.) Matsum. & Nakai]. Theor Appl Genet 125:1603–1618
Shang J, Li N, Li N, Xu Y, Ma S, Wang J (2016) Construction of a high-density genetic map for watermelon (Citrullus lanatus L.) based on large-scale SNP discovery by specific length amplified fragment sequencing (SLAF-seq). Sci Hort 203:38–46
Singh SJ, Krishnareddy M (1996) Watermelon bud necrosis: new Tospovirus disease. Acta Hort 431:68–77
Stevens MR, Lamb EM, Rhoads DD (1995) Mapping the Sw-5 locus for tomato spotted wilt virus resistance in tomatoes using RAPD and RFLP analyses. Theor Appl Genet 90:451–456
Sugiyama M, Kawazu Y, Fukino N, Yoshioka Y, Shimomura K, Sakata Y, Okuda M (2015) Mapping of quantitative trait loci for Melon yellow spot virus resistance in cucumber (Cucumis sativus L.). Euphytica 205:615–625
Sugiyama M, Okuda M, Sakata Y (2009) Evaluation of resistance to melon yellow spot virus in a cucumber germplasm collection. Plant Breeding 128:696–700
Sun X, Mumm RH (2016) Method to represent the distribution of QTL additive and dominance effects associated with quantitative traits in computer simulation. BMC Bioinform 17:73. https://doi.org/10.1186/s12859-016-0906-z
Wang C, Qiao A, Fang X, Sun L, Peng P, Davis AR, Liu S, Luan F (2019) Fine mapping of lycopene content and flesh color related gene and development of molecular marker-assisted selection for flesh color in watermelon (Citrullus lanatus). Front Plant Sci 10:1240. https://doi.org/10.3389/fpls.2019.01240
Wang J, Li H, Zhang L, Meng L (2016) Users manual of QTL IciMapping. The Quantitative Genetics Group, Institute of Crop Science, Chinese Academy of Agricultural Sciences (CAAS), Beijing 100081, China, and Genetic Resources Program, International Maize and Wheat Improvement Center (CIMMYT), Apdo. Postal 6-641, 06600 Mexico, D.F., Mexico, p 274
Wu S, Wang X, Reddy U, Sun H, Bao K, Gao L, Mao L, Patel T, Ortiz C, Abburi V, Nimmakayala P, Branham S, Wechter P, Massey L, Ling KS, Kousik C, Hammar S, Tadmor Y, Portnoy V, Fei Z (2019) Genome of ‘Charleston Gray’, the principal American watermelon cultivar, and genetic characterization of 1365 accessions in the U. S. National Plant Germplasm System watermelon collection. Plant Biotech J 17(12):2246–2258
Zhang H, Wang H, Guo S, Ren Y, Gong G, Weng Y, Xu Y (2012) Identification and validation of a core set of microsatellite markers for genetic diversity analysis in watermelon (Citrullus lanatus Thunb. Matsum Nakai). Euphytica 186(2):329–342
Zhang R, Xu Y, Yi K, Zhang H, Liu L, Gong G, Levi A (2004) A genetic linkage map for watermelon derived from recombinant inbred lines. J American Soc Hort Sci 129:237–324
Zhang Z, Zhang Y, Sun L, Qiu G, Sun Y, Zhu Z, Luan F, Wang X (2018) Construction of a genetic map for Citrullus lanatus based on CAPS markers and mapping of three qualitative traits. Sci Hort 233:532–538
Acknowledgements
The authors would like to acknowledge Ministry of Social Justice and Empowerment, Government of India, for providing the Ph.D fellowship for first author and Department of Biotechnology, Government of India, for funding the program.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: ESR; NGC, TRN, SKM and MR performed the experiments with assistance from MKR for phenotyping; LRDC for genotyping; AR for viral diagnostic assays; ESR and NGC wrote the paper. All the authors have discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nagesh, G.C., Thontadarya, R.N., Swamy, K.M. et al. Mapping quantitative trait loci for resistance to watermelon bud necrosis orthotospovirus in watermelon [Citrullus lanatus (Thunb.) Matsum & Nakai]. Euphytica 216, 104 (2020). https://doi.org/10.1007/s10681-020-02632-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-020-02632-8