Abstract
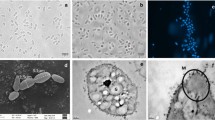
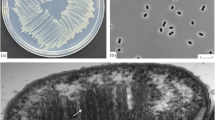
A novel gammaproteobacterial methanotroph; strain FWC3 was isolated from a tropical freshwater wetland sample collected near a beach in Western India. Strain FWC3 forms flesh pink/peach colored colonies, is non-motile, and the cells are present as diplococci, triads, tetracocci and aggregates. Strain FWC3 grows only on methane and methanol. As the 16S rRNA gene of strain FWC3 showed low similarities with other Type I methanotrophs (less than 94.3%), it was further investigated for its novelty and characterisation by a polyphasic approach. ANI indices and DDH values deduced from the draft genome of strain FWC3 (SEYW00000000.1) with the other nearest type strains (Methylocaldum marinum S8T and Methylococcus capsulatus BathT) were ~ 70% and ~ 15%, respectively. The low level similarities indicated that strain FWC3 can belong to a new genus and species. Additionally, strain FWC3 showed a unique fatty acid profile with the dominance of C16:1 ω7 and ω6c, C16:0 and C16:1 ω9c. During the characterisation of strain FWC3, a morphologically similar methanotroph, strain C50C1 was described (Ghashghavi et al. in mSphere 4:e00631–18, 2019) and named as ‘Methylotetracoccus oryzae’. We found that strain FWC3 and strain C50C1 belonged to the same genus but could belong to different species based on the ANI indices and dDDH values (~ 94% and ~ 55%, respectively). However, strain C50C1 has not been deposited in two culture collections and not been validly described. Also, the 16S rRNA gene of strain C50C1 is neither available on the database nor can it be retrieved from the genome assembly. Based on the polyphasic characterisation and comparison to the other type strains of Methylococcaceae, we propose strain FWC3 (= JCM 33786T, = KCTC 72733T, = MCC 4198T) to be the type strain of a novel genus and species, for which the name Methylolobus aquaticus is proposed. Strain C50C1 (Ghashghavi et al. 2019) could represent another species (‘Methylolobus oryzae’).



Similar content being viewed by others
References
Chun J, Oren A, Ventosa A, Christensen H, Araha DR, da Costa MS et al (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Conrad R (2009) The global methane cycle: recent advances in understanding the microbial processes involved. Environ Microbiol Rep 1:285–292
Dedysh SN, Knief C (2018) Diversity and phylogeny of described aerobic methanotrophs. In: Kalyuzhnaya MG, Xing X-H (eds) Methane biocatalysis: paving the way to sustainability. Springer, New York
Frindte K, Maarastawi SA, Lipski A, Hamacher J, Knief C (2017) Characterization of the first rice paddy cluster I isolate, Methyloterricola oryzae gen. nov., sp. nov. and amended description of Methylomagnum ishizawai. Int J Syst Evol Microbiol 67:4507–4514
Geymonat E, Ferrando L, Tarlera SE (2011) Methylogaea oryzae gen. nov., sp. nov., a mesophilic methanotroph isolated from a rice paddy field. Int J Syst Evol Microbiol 61:2568–2572
Ghashghavi M, Belova SE, Bodelier PLE, Dedysh SN, Kox MAR, Speth DR, Frenzel P, Jetten MSM, Lücker S, Lüke C (2019) Methylotetracoccus oryzae strain C50C1 Is a novel Type Ib gammaproteobacterial methanotroph adapted to freshwater environments. mSphere 4:e00631–18
Hanson RS, Hanson TE (1996) Methanotrophic bacteria. Microbiol Mol Biol Rev 60:439–471
Khatri K, Pandit P, Mohite J, Bahulikar R, Rahalkar M (2019a) A novel gammaproteobacterial methanotroph from Methylococccaeae; strain FWC3, isolated from canal sediment from Western India. BioRxiv. https://doi.org/10.1101/696807
Khatri K, Mohite JA, Pandit PS, Bahulikar RA, Rahalkar MC (2019b) Description of ‘Ca. Methylobacter oryzae’ KRF1, a novel species from the environmentally important Methylobacter clade 2. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01369-2(0123456789)
Knief C (2015) Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front Microbiol 6:1346
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 60:1–14
Oren A, Garrity G (2019) List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 69:2627–2629
Pandit P, Rahalkar MC (2018) Renaming of ‘Candidatus Methylocucumis oryzae’ as Methylocucumis oryzae gen. nov., sp. nov., a novel Type I methanotroph isolated from India. Antonie Van Leeuwenhoek 112:955–959
Pandit PS, Rahalkar MC, Dhakephalkar PK, Ranade DR, Pore S, Arora P, Kapse N (2016) Deciphering community structure of methanotrophs dwelling in rice rhizospheres of an Indian rice field using cultivation and cultivation-independent approaches. Microb Ecol 71:634–644
Pandit PS, Hoppert M, Rahalkar MC (2018) Description of ‘Candidatus Methylocucumis oryzae’, a novel Type I methanotroph with large cells and pale pink colour, isolated from an Indian rice field. Antonie Van Leeuwenhoek 111:2473–2484
Rahalkar MC, Pandit PS (2018) Genome-based insights into a putative novel Methylomonas species (strain Kb3), isolated from an Indian rice field. Gene Rep 13:9–13
Rahalkar MC, Pandit PS, Dhakephalkar PK, Pore S, Arora P, Kapse N (2016) Genome characteristics of a novel Type I methanotroph ‘Sn10-6’ isolated from a flooded Indian rice field. Microb Ecol 71:519–523
Rahalkar MC, Khatri K, Pandit PS, Dhakephalkar PK (2019) A putative novel Methylobacter member (KRF1) from the globally important Methylobacter clade 2: cultivation and salient draft genome features. Antonie Van Leeuwenhoek 112:1399–1408
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131
Sangal V, Goodfellow M, Jones AL, Schwalbe EC, Blom J, Hoskisson PA, Sutcliffe IC (2016) Next-generation systematics: an innovative approach to resolve the structure of complex prokaryotic taxa. Sci Rep 6:1–12
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, Gerdes S, Henry RW, Kenyon D, Machi C, Mao EK, Nordberg GJ, Olsen DE, Murphy-Olson R, Olson R, Overbeek B, Parrello GD, Pusch SM, Vonstein V, Warren A, Xia F, Yoo H, Stevens RL (2017) Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acid Res 45:D535–D542
Acknowledgements
We thank Ms. Ambika Pote for her assistance during some experiments. We also thank Agharkar Research Institute for the overall support. We thank Mr. Atul Dviwedi for the technical assistance during SEM. We are very thankful to Prof. Bernhard Schink for his help in the etymology. We are thankful to the three culture collections in this study (NCMR, Pune, KCTC, Korea and JCM, Japan) for the cultivation and preservation of the strain.
Funding
MCR acknowledges Department of Science and Technology, DST, SERB (EMR/2017/002817) for the funds. PSP acknowledges DST (SR/WOS-A/LS-410/2017) for providing the women scientist fellowship funds. Kumal Khatri acknowledges CSIR for providing junior research and senior research fellowship.
Author information
Authors and Affiliations
Contributions
Conceptualization: MCR and RAB. Data Analysis: MCR, RAB and KK. Experimental work: Strain isolation, characterisation, maintenance and growth experiments: KK, JM and PSP. Fund acquisition: MCR. Phylogenetic analysis and sample collection: MCR and RAB. Writing of the manuscript: MCR. Writing-review and editing: MCR and RAB. Final version reviewed and approved by all the authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rahalkar, M.C., Khatri, K., Mohite, J. et al. A novel Type I methanotroph Methylolobus aquaticus gen. nov. sp. nov. isolated from a tropical wetland. Antonie van Leeuwenhoek 113, 959–971 (2020). https://doi.org/10.1007/s10482-020-01410-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-020-01410-9