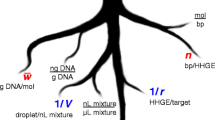
Abstract
Digital polymerase chain reaction (dPCR) methodology has been asserted to be a “potentially primary” analytical approach for assigning DNA concentration. The essence of dPCR measurements is the independent dispersal of fragments into multiple reaction partitions, amplifying fragments containing a target nucleotide sequence until the signal from all partitions containing at least one such fragment rises above threshold, and then determining the proportion of partitions with an above-threshold signal. Should originally double-stranded DNA (dsDNA) fragments be converted into two single strands (ssDNA) prior to dispersal, the dPCR measurements could be biased high by as much as a factor of two. Realizing dPCR’s metrological potential therefore requires analytical methods for determining the proportion of ssDNA in nominally dsDNA samples. To meet this need, we have investigated several approaches to this determination: A260 ratio, dPCR ratio, cdPCR staircase, and ddPCR enzyme. In our hands, only the endonuclease-based approach provides adequately accurate estimates for relatively small ssDNA proportions. We present four (enzyme, assay) pairs that provide self-consistent results for human nuclear DNA extracts. However, the proportion of ssDNA differs by as much as 50% between assays, apparently related to the guanine-cytosine (GC) content of the fragment near the assay’s target sequence. While materials extracted by us have no more than 6% ssDNA content even after long storage, a commercially obtained PCR assay calibrant contains ≈18% ssDNA.

Graphical abstract









Similar content being viewed by others
Data availability
The summary ddPCR and cdPCR data used are presented as Tables S1 and S2 in the ESM. The aA, aB, and aC materials are components A, B, and C of SRM 2372a and are available for purchase from NIST through https://www.nist.gov/srm. The spreadsheet-based cdPCR Staircase analysis system is available on request from the corresponding author.
References
Kline MC, Duewer DL. Evaluating digital polymerase chain reaction for the quantification of human genomic DNA: lifting the traceability fog. Anal Chem. 2017;89(8):4648–54. https://doi.org/10.1021/acs.analchem.7b00240.
Dagata JA, Farkas N, Kramer JA. Method for measuring the volume of nominally 100 μm diameter spherical water-in-oil emulsion droplets. NIST Special Publication (NIST SP) 260–184. 2016. https://doi.org/10.6028/NIST.SP.260-184.
Duewer DL, Kline MC, Romsos EL, Toman B. Evaluating droplet digital PCR for the quantification of human genomic DNA: converting copies per nanoliter to nanograms nuclear DNA per microliter. Anal Bioanal Chem. 2018;410(12):2879–87. https://doi.org/10.1007/s00216-018-0982-1.
Romsos EL, Kline MC, Duewer DL, Toman B, Farkas N. Certification of standard reference Material® 2372a human DNA quantitation standard. NIST Special Publication (NIST SP) 260-189, 2018. https://doi.org/10.6028/NIST.SP.260-189.
NIST certificates of analysis are available through the NIST standard reference materials homepage. https://www.nist.gov/srm. Accessed 26 May 2020.
Bhat S, Curach N, Mostyn T, Bains GS, Griffiths KR, Emslie KR. Comparison of methods for accurate quantification of DNA mass concentration with traceability to the international system of units. Anal Chem. 2010;82:7185–92. https://doi.org/10.1021/ac100845m.
Sanders R, Huggett JF, Bushell CA, Cowen S, Scott DJ, Foy CA. Evaluation of digital PCR for absolute DNA quantification. Anal Chem. 2011;83:6474–84. https://doi.org/10.1021/ac103230c.
Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001.
ISO 21571:2005; Foodstuffs – Methods of Analysis for the Detection of Genetically Modified Organisms and Derived Products – Nucleic Acid Extraction, Annex B Methods for Quantitation of Extracted DNA; International Standards Organization: Geneva, Switzerland: 2005, pp. 34–36.
Wilson PJ, Ellison SLR. Extending digital PCR analysis by modelling quantification cycle data. BMC Bioinformatics. 2016;17:421. https://doi.org/10.1186/s12859-016-1275-3.
Kline MC, Duewer DL. Evaluation of methods for assessing the proportion of single stranded nuclear DNA in human blood extracts. NIST Special Publication (NIST SP) 1200-27, 2019. https://doi.org/10.6028/NIST.SP.1200-27.
Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. https://doi.org/10.1093/nar/16.3.1215.
Cavaluzzi MJ, Borer PN. Revised UV extinction coefficients for nucleoside-5′-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004;32(1):e13. https://doi.org/10.1093/nar/gnh015.
Nwokeoji AO, Kilby PM, Portwood DE, Dickman MJ. Accurate quantification of nucleic acids using hypochromicity measurements in conjunction with UV spectrophotometry. Anal Chem. 2017;89(24):13567–74. https://doi.org/10.1021/acs.analchem.7b04000.
Volkov SN, Danilov VI. Study of 1st and 2nd absorption-band hypochromism in natural DNA. FEBS Lett. 1976;65(1):8–10. https://doi.org/10.1016/0014-5793(76)80609-6.
Tinoco I. Hypochromism in polynucleotides. J Am Chem Soc. 1960;82(18):4785–90. https://doi.org/10.1021/ja01503a007.
Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon JL, Morley A. Quantitation of targets for PCR by use of limiting dilution. BioTechniques. 1992;13(3):444–9.
Duewer DL, Kline MC, Romsos EL. Real-time cdPCR opens a window into events occurring in the first few PCR amplification cycles. Anal Bioanal Chem. 2015;407(30):9061–9. https://doi.org/10.1007/s00216-015-9073-8.
Kline MC, Romsos EL, Duewer DL. Evaluating digital PCR for the quantification of human genomic DNA: accessible amplifiable targets. Anal Chem. 2016;88(4):2132–9. https://doi.org/10.1021/acs.analchem.5b03692.
Zhou Y, Paull TT. Direct measurement of single-stranded DNA intermediates in mammalian cells by quantitative polymerase chain reaction. Anal Biochem. 2015;479:48–50. https://doi.org/10.1016/j.ab.2015.03.025.
Nishigaki K, Kaneko Y, Wakuda H, Husimi Y, Tanaka T. Type II restriction endonucleases cleave single-stranded DNAs in general. Nucleic Acids Res. 1985;13(16):5747–60.
Thompson M. Uncertainty functions, a compact way of summarising or specifying the behaviour of analytical systems. TrAC. 2011;30(7):1168–75. https://doi.org/10.1016/j.trac.2011.03.012.
Bikard D, Loot C, Baharoglu Z, Mazel D. Folded DNA in action: hairpin formation and biological functions in prokaryotes. Microbiol Mol Biol Rev. 2010;74(4):570–88. https://doi.org/10.1128/MMBR.00026-10.
Ripley BD, Thompson M. Regression techniques for analytical bias. Analyst. 1987;112:377–83. https://doi.org/10.1039/AN9871200377.
Analytical Methods Committee. Linear functional relationship estimation by maximum likelihood. https://www.rsc.org/Membership/Networking/InterestGroups/Analytical/AMC/Software/FREML.asp. Accessed 26 May 2020.
Yakovchuk P, Protozanova E, Frank-Kamenetskii MD. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006;34(2):564–74. https://doi.org/10.1093/nar/gkj454.
Magnusson B, Ellison SLR. Treatment of uncorrected measurement bias in uncertainty estimation for chemical measurements. Anal Bioanal Chem. 2008;390(1):201–13. https://doi.org/10.1007/s00216-007-1693-1.
Funding
This work was supported in part by the NIST Special Programs Office project Forensic DNA.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest/Competing interests
The authors declare that they have no conflict of interest nor competing interests.
Ethics approval
All work presented has been reviewed and approved by the National Institute of Standards and Technology Human Subjects Protections Office. This study was determined to be “not human subjects research” (often referred to as research not involving human subjects) as defined in U. S. Department of Commerce Regulations, 15 CFR 27, also known as the Common Rule (45 CFR 46, Subpart A), for the Protection of Human Subjects.by the NIST Human Subjects Protection Office and therefore not subject to oversight by the NIST Institutional Review Board.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
DISCLAIMER: Certain commercial equipment, instruments, or materials are identified in this report to specify adequately experimental conditions or reported results. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the equipment, instruments, or materials identified are necessarily the best available for the purpose.
Electronic supplementary material
ESM 1
(PDF 812 kb)
Rights and permissions
About this article
Cite this article
Kline, M.C., Duewer, D.L. Evaluating digital PCR for the quantification of human nuclear DNA: determining target strandedness. Anal Bioanal Chem 412, 4749–4760 (2020). https://doi.org/10.1007/s00216-020-02733-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-020-02733-2