Abstract
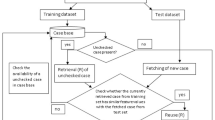
Medical image processing is now gaining a significant momentum in clinical situation to undertake diagnosis of different anatomical defects. However, with regard to eye diseases, there is no such well-defined image processing technique in medical image analysis. The scope of this study is to automate computer analysis of ocular disease-related retinal images, which may ease the job of ophthalmologists to rule out the diseased condition. In this present work, eye images are subjected for developing a reliable tool for processing the eye retinal fundus images. The primary objective is to effectively probe retinal image data for providing a holistic approach in automatic fundus disease detection and screening to help clinicians in addition with a developed reliable image processing technique combined with a rule-based clustering method for automatic analysis of fundus images in a reduced time frame. More than 400 eye images available in online are examined. The images were preprocessed by grayscale conversion, retinal segmentation, ROI and crop ROI, image resizing, and extraction in RGB channels. Then these images were segmented by NRR from RGB channels, centroids of rows and columns, and NRR to binary image conversion. Then extraction of features like cup to disc area, optic cup area, and NRR calculations prior to measuring ISNT. A unique algorithm named as EARMAM was introduced for the prediction of diseased image from healthy eye image pool is envisaged in this paper. The functional significance of the EARMAM algorithm was compared with other common classification algorithm of current practice such as SVM, naïve Bayes, random forest, and SMO. The results of confusion matrix have shown that there was 93% prediction accuracy which was higher than the predictive values of other algorithms. The above results are discussed with future improvement and application in clinical field.









Similar content being viewed by others
References
Abramoff MD, Niemeijer M, Suttorp-Schulten MSA, Viegever MA, Russel SR, van Ginneken B (2008) Evaluation of a system for automatic detection of diabetic retinopathy from color fundus photographs in a large population of patients with diabetes. Diabetes Care 31:193–197. https://doi.org/10.2337/dc07-1312
Mitsuishi Y, Motohashi H, Yamamoto M (2012) The Keap1-Nrf2 system in cancers: stress response and anabolic metabolism. Front Oncol 2:200. https://doi.org/10.3389/fonc.2012.00200
McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD (2004) Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J Biol Chem 279(30):31556–31567. https://doi.org/10.1074/jbc.M403061200
Ho Kim J, Franck J, Kang T, Heinsen H, Ravid R, Ferrer I, Hee Cheon M, Lee JY, Shin Yoo J, Steinbusch HW, Salzet M, Fournier I, Mok Park Y (2015) Proteome-wide characterization of signalling interactions in the hippocampal CA4/DG subfield of patients with Alzheimer’s disease. Sci Rep 5:11138. https://doi.org/10.1038/srep11138
Miki Y, Tanji K, Mori F, Kakita A, Takahashi H, Wakabayashi K (2017) Alteration of mitochondrial protein PDHA1 in Lewy body disease and PARK14. Biochem Bioph Res Co 489(4):439–444
Gokcal E, Gur VE, Selvitop R, Babacan Yildiz G, Asil T (2017) Motor and non-motor symptoms in parkinson’s disease: effects on quality of life. Noro Psikiyatri Arsivi 54(2):143–148. https://doi.org/10.5152/npa.2016.12758
Heng X, Zhu BJJ, Sun LM, Zhang QC (2018) Network pharmacology-based study on mechanisms of Huanglian Jiedu Decoction impact on macrophage inflammation response. Acta Pharm Sin 53(9):1449–14457. https://doi.org/10.16438/j.0513-4870.2018-0276
Grondin CJ, Davis AP, Wiegers TC, Wiegers JA, Mattingly CJ (2018) Accessing an expanded exposure science module at the comparative toxicogenomics database. Environ Health Perspect 126(1):014501. https://doi.org/10.1289/EHP2873
Liu Y, Guo Y, Wu W et al (2019) A machine learning-based QSAR model for benzimidazole derivatives as corrosion inhibitors by incorporating comprehensive feature selection. Interdiscip Sci Comput Life Sci 11:738–747. https://doi.org/10.1007/s12539-019-00346-7
Yang F, Diao X, Wang F et al (2020) Identification of key regulatory genes and pathways in prefrontal cortex of Alzheimer’s disease. Interdiscip Sci Comput Life Sci 12:90–98. https://doi.org/10.1007/s12539-019-00353-8
AcharyaI UR (2011) Automated diagnosis of glaucoma using texture and higher order spectra features. IEEE Trans Inform Technol Biomed 15(3):449–455
Huang ML, Chen HY, Huan JJ (2007) Glaucoma detection using adaptive neuro-fuzzy inference system. Expert Syst Appl 32(2):458–468
Hu B, Chang X, Liu X (2019) Predicting functional modules of liver cancer based on differential network analysis. Interdiscip Sci Comput Life Sci 11:636–644. https://doi.org/10.1007/s12539-018-0314-3
Miao YR, Liu W, Zhang Q et al (2018) lncRNASNP2: an updated database of functional SNPs and mutations in human and mouse lncRNAs. Nucleic Acids Res 46:D276–D280
Saumya M, Subin EK, Suchithra TV (2019) Network analysis of MPO and other relevant proteins involved in diabetic foot ulcer and other diabetic complications. Interdiscip Sci Comput Life Sci 11:180–190. https://doi.org/10.1007/s12539-017-0258-z
Suresh A, Shunmuganathan KL (2012) Image texture classification using gray level co-occurrence matrix based statistical features. Eur J Sci Res 75(4):591–597 (ISSN 1450-216X)
Suresh A, Udendhran R, Balamurgan M et al (2019) A novel internet of things framework integrated with real time monitoring for intelligent Healthcare environment. J Med Syst 43:165. https://doi.org/10.1007/s10916-019-1302-9
Liu D, Kong Z, Wang Y et al (2019) Quantitative and visual characteristics of primary central nervous system lymphoma on 18F-FDG-PET. Interdiscip Sci Comput Life Sci 11:300–306. https://doi.org/10.1007/s12539-019-00333-y
Li B, Yang F (2018) An across-target study on visual attentions in facial expression recognition. Interdiscip Sci Comput Life Sci 10:367–374. https://doi.org/10.1007/s12539-018-0281-8
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
In accordance with all authors, we are reporting that there is no conflict of interest in the research paper.
Ethical Responsibilities of Authors
The authors follow the ethical information provided in the journal and hereby abide the same with the journal.
Rights and permissions
About this article
Cite this article
Karthiyayini, R., Shenbagavadivu, N. Retinal Image Analysis for Ocular Disease Prediction Using Rule Mining Algorithms. Interdiscip Sci Comput Life Sci 13, 451–462 (2021). https://doi.org/10.1007/s12539-020-00373-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-020-00373-9