Abstract
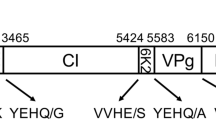
We determined the complete genomic sequence of begonia flower breaking virus (BFBV), a novel putative member of the genus Potyvirus isolated from Begonia bowerae cv. ‘Tiger’ plants grown in Kunming. The genomic RNA comprises 9540 nucleotides (nt), excluding the 3’-terminal poly(A) tail, and contains a typical open reading frame (ORF) of potyviruses. The ORF consists of 9219 nucleotides and encodes a 3073-amino-acid polyprotein that is predicted to be proteolytically cleaved into 10 mature peptides. Sequence comparison indicated that BFBV shares 43.9–55.12% amino acid sequence identity with known potyviruses and that BFBV shares the highest amino acid sequence identity (55.12%) with beet mosaic virus. The results from the complete genomic sequence analysis further suggest that BFBV is a member of a novel species in the genus Potyvirus.


Similar content being viewed by others
References
Bouwen R, Vlugt VD (2005) A new potyvirus causing flower breaking in Begonia semperflorens. Fourth Joint Meeting of Dutch and German Plant Virologists 21
Wu HZ, Gao J, Xu Y, Wu JZ, He SL, Cheng HR (2016) First report of begonia flower breaking virus infecting begonia bowerae in China. Plant Dis 100:2538
Dey KK, Li C, Douglas K, McVay J, Whilby L, Hodges G, Smith TR (2019) First report of Begonia flower breaking virus Infecting Begonia (Begonia spp.) in the USA. Plant Dis https://www.researchgate.net/publication/335474771
Zheng Y, Gao S, Padmanabhan C, Li R, Fei Z (2017) Virusdetect: an automated pipeline for efficient virus discovery using deep sequencing of small RNAs. Virology 500:130–138
Xu DL, Liu HY, Koike ST, Li F, Li R (2011) Biological characterization and complete genomic sequence of Apium virus Y infecting celery. Virus Res 155(1):82
Vlugt VD, Steffens P, Cuperus C, Barg E, Vetten HJ (1999) Further evidence that shallot yellow stripe virus (SYSV) is a distinct potyvirus and reidentification of welsh onion yellow stripe virus as a sysv strain. Phytopathology 89(2):148–155
Adams MJ, Antoniw JF, Beaudoin F (2005) Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol Plant Pathol 6:471–487
Olspert A, Chung BY, Atkins JF, Carr JP, Firth AE (2015) Transcriptional slippage in the positive-sense RNA virus family Potyviridae. EMBO Rep 16:995–1004
Shiboleth YM, Haronsky E, Leibman D, Arazi T, Wassenegger M, Whitham SA, Gaba V, Gal-On A (2007) The conserved FRNK box in HC-Pro, a plant viral suppressor of gene silencing, is required for small RNA binding and mediates symptom development. J Virol 81:13135–13148
Urcuqui-Inchima S, Haenni AL, Bernardi F (2001) Potyvirus proteins: a wealth of functions. Virus Res 74:157–1755
Kumar S, Stecher G, Tamura K (2016) Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870
Wylie SJ, Adams M, Chalam C, Kreuze J, Lopez-Moya JJ et al (2017) ICTV virus taxonomy profile: potyviridae. J Gen Virol 98:352–354
Funding
This study was funded by the Project for High-Level Creative University Students in Horticulture from the Educational Department of Yunnan (2017CX015) and the Academician Expert Workstation from the Yunnan Provincial Science and Technology Department (2018IC098).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
As the corresponding author, Hongzhi Wu declares that no conflicts of interest are involved with the authors of this paper.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Stephen John Wylie.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tang, Y., Wu, J., Zhao, M. et al. Complete genome sequence of begonia flower breaking virus, a novel member of the genus Potyvirus. Arch Virol 165, 1915–1918 (2020). https://doi.org/10.1007/s00705-020-04663-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04663-x