Abstract
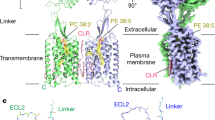
Metabotropic GABAB G protein-coupled receptor functions as a mandatory heterodimer of GB1 and GB2 subunits and mediates inhibitory neurotransmission in the central nervous system. Each subunit is composed of the extracellular Venus flytrap (VFT) domain and transmembrane (TM) domain. Here we present cryo-EM structures of full-length human heterodimeric GABAB receptor in the antagonist-bound inactive state and in the active state complexed with an agonist and a positive allosteric modulator in the presence of Gi1 protein at a resolution range of 2.8–3.0 Å. Our structures reveal that agonist binding stabilizes the closure of GB1 VFT, which in turn triggers a rearrangement of TM interfaces between the two subunits from TM3-TM5/TM3-TM5 in the inactive state to TM6/TM6 in the active state and finally induces the opening of intracellular loop 3 and synergistic shifting of TM3, 4 and 5 helices in GB2 TM domain to accommodate the α5-helix of Gi1. We also observed that the positive allosteric modulator anchors at the dimeric interface of TM domains. These results provide a structural framework for understanding class C GPCR activation and a rational template for allosteric modulator design targeting the dimeric interface of GABAB receptor.
Similar content being viewed by others
Introduction
Metabotropic GABAB receptor is a G protein-coupled receptor (GPCR) for the major inhibitory neurotransmitter, γ-amino butyric acid (GABA), in the central nervous system, which mediates slow and prolonged inhibitory activity.1,2,3 GABAB receptor couples to Gi/o proteins to inhibit neurotransmitter release in pre-synaptic neurons and cause hyperpolarization in the post-synaptic neurons.4 GABAB receptor suppresses adenylyl cyclase through Gɑi/o, while gating ion channels and transactivating receptor tyrosine kinases via Gβγ.5,6,7,8 Dysfunctions of GABAB receptor or mutation in its genes have been implicated in a variety of neurological and psychiatric disorders including epilepsy, pain, anxiety, depression, schizophrenia, drug addiction, Rett syndrome and epileptic encephalopathies.4,9 Recent studies reveal that the auto-antibodies of GABAB receptor are possibly the origin of epilepsies and encephalitis.10,11 Although a large number of antagonists and agonists and positive or negative allosteric modulators (PAMs or NAMs) have been developed for GABAB receptor,12 only two agonists have been approved as therapeutic drugs: baclofen (Lioresal®) used for the treatment of muscle spasticity and alcohol addiction13,14 and γ-hydroxybutyrate (GHB) used for the treatment of narcolepsy.15
GABAB receptor belongs to the class C GPCR composed of metabotropic glutamate receptors (mGlus), calcium-sensing receptor (CaSR) and taste 1 receptors.16 Class C GPCR are obligatory dimers; and among them, mGlus and CaSR are the homodimers,17,18 whereas GABAB receptor is an obligatory heterodimer of two subunits, GABAB1 (GB1) and GABAB2 (GB2).19,20 Each subunit is composed of a large extracellular “Venus Flytrap” (VFT) domain, a transmembrane (TM) domain, and a cytoplasmic tail21 (Supplementary information, Fig. S1a, b). GB1 possesses an endoplasmic reticulum (ER) retention motif in its cytoplasmic tail.22 The interaction between GB1 and GB2 facilitates its cell surface expression through coiled-coil interactions in its cytoplasmic tail.23 While GB1 is responsible for ligand recognition through its VFT,24 GB2 couples Gi/o proteins through its TM.25,26 However, a line of evidence shows that in the absence of GB2, GB1asa (a GB1 mutant with deletion of ER retention signal) at the cell surface or ER-localized GB1 also induces downstream signaling through Gi/o proteins.27,28
The crystal structures of isolated heterodimeric VFT of GABAB receptor in the presence of antagonist or agonist revealed the open or closed conformation of GB1 VFT.29 Structures of isolated VFT and TM domains from other class C GPCR have been reported,30,31,32 in addition to the more recent breakthrough that cryo-EM structures of full-length mGlu5 in apo state and in the presence of agonist have been determined both at overall 4 Å resolution, providing the first insights into the architecture of mGlu5 and structural framework of mGlu5 activation.33 However, no structure of full-length GABAB receptor has been solved, limiting our understanding of the configuration of GABAB receptor. Moreover, molecular mechanism of class C GPCR signal transduction remains elusive, primarily owning to the lack of structural information of the receptor in various states, especially coupled to a downstream G protein. Here we present cryo-EM structures of full-length human heterodimeric GABAB receptor in the antagonist-bound inactive state and in the agonist/PAM-bound active state complexed with heterotrimeric Gi1 protein, shedding lights on class C GPCR activation and providing a rational template for allosteric modulator design targeting the dimeric interface of GABAB receptor.
Results
Cryo-EM structure determination of GABAB receptor
We first sought to obtain the heterodimeric human GABAB receptor in the fully inactive state. Thus, we overexpressed both GB1a and GB2 subunits in full length in mammalian cells and maintained 20 μM CGP54626, a potent antagonist with estimated nano-molar affinity to the receptor,34 through the sample preparation stage. The heterodimeric GABAB receptor was solubilized from the membrane using detergent. The following visualization by negative-staining EM displayed the existence of the intact heterodimer, and unlike detergent-reconstituted mGlu5 in the inactive (apo) state with split micelles,33 GABAB receptor showed one large disc-shaped micelle containing TMs from both subunits (Supplementary information, Fig. S1c, d). Two-dimensional class averages of frozen specimen showed clear secondary structure features for GABAB receptor (Supplementary information, Fig. S2); this confirmed that two TM bundles were embedded in the detergent micelle suggesting the more stable intersubunit interactions between TM domains of GABAB receptor compared to that of mGlu5.33 We therefore obtained cryo-EM images and determined the density map for CGP54626-bound GABAB receptor using single-particle cryo-EM to an overall resolution of 3.0 Å. Local resolution calculations indicate a range of 2.4–3.4 Å in most map regions (Fig. 1a and Supplementary information, Fig. S2). The antagonist, VFT and TM domains from both subunits are clearly visible in the cryo-EM map (Fig. 1a), except the sushi domains at GB1 N-terminus and coiled-coil domain at the GABAB receptor cytoplasmic tail, indicating the dynamic nature of these two regions.
a, b Cryo-EM density maps (left), models (middle) and maps colored according to local resolution (right) for the GABAB receptor in the presence of CGP54626 (slate; antagonist) (a) and in complex with baclofen (magenta; agonist), BHFF (steel blue; PAM) and Gi1 protein (b). The colored density map is a composite map generated with the VFT and TMD locally refined map. The cryo-EM density map before focused refinement in transparent superposed with the final map, illustrating density for detergent micelle and Gi1 protein. The local resolution (Å) was calculated with the locally refined map as input, indicating a range of 2.5–3.5 Å resolution in most map regions for both the inactive and active states. GB1 and GB2 in inactive state, blue and yellow, respectively; red and green in active state, respectively.
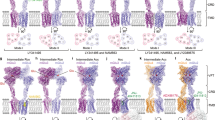
To prepare the stable and sufficient activated GABAB–Gi1 complex for cryo-EM study, initial efforts were made to accomplish the complex formation under the condition in which the purified receptor and the Gi1 protein were incubated with baclofen, a commercially available PAM BHFF in racemate form (rac-BHFF)35 and Gi-binding protein scFv16,36 unfortunately leading to the aggregation of the complex (data not shown). Considering the flexibility of the coiled-coil domain indicated by the inactive structure of the full-length receptor, we thus removed it and the pull-down assay suggested that the receptor lacking the coiled-coil domain (GABABΔcc) exhibited increased expression level therefore leading to the improved yield of GABAB–Gi1 complex (Supplementary information, Fig. S1e). Previous electrophysiological studies showed that these truncations of the receptor did not attenuate its ability to activate the inwardly rectifying K+ current;37 our fluorescence assay also suggested that these modifications do not alter receptor pharmacology (hereafter referring to GABABΔcc as GABAB unless otherwise noted) (Supplementary information, Fig. S1f). We therefore formed the baclofen/PAM–GABAB–Gi1 complex, which was solubilized using lauryl maltoseneopentyl glycol and purified by tandem affinity and size-exclusion chromatography (Supplementary information, Fig. S1g). Sample evaluation by negative-stain EM confirmed the presence of Gi1 protein and monodispersed particle distribution (Supplementary information, Fig. S1h). In addition, cryo-EM analysis suggested multiple conformations to be spontaneously present in the GABAB–Gi1 complex and three-dimensional classification revealed three major conformers, with one of them representing Gi1 protein engaged with GB1 and the other two representing Gi1 protein coupled to GB2 in distinct orientations (termed B1, B2a and B2b state, respectively). We further refined these three conformers independently and obtained the reconstructions with nominal resolutions of 8.8 Å, 6.8 Å and 8.6 Å, respectively, which were adequate for rigid-body docking of the receptor and Gi1 independently to generate models for their relative arrangement (Supplementary information, Fig. S3). Close visualization and superimposition of three conformers with each other revealed no distinguishable difference in the receptor part, enabling us to obtain high-resolution reconstruction for the active receptor (Supplementary information, Figs. S3, S4). Therefore, we focused particle projection alignment on the VFT module and TM domain of the receptor separately to improve the map quality and managed to obtain the maps with the resolutions of 2.9 Å and 3.0 Å, respectively, which were subsequently combined to produce a composite map for model building and refinement (Fig. 1b and Supplementary information, Fig. S3).
Taken together, the high-quality density maps allow the unambiguous placement of antagonist (CGP54626), agonist (baclofen), PAM (BHFF) and most side-chain amino acids of the receptor in both inactive and active states including all the extracellular loops (ECLs) and intracellular loops (ICLs) except ICL2 (Fig. 3e; Supplementary information, Figs. S5, S7). Thus, our structures provide accurate models of the full-length GABAB receptor in the inactive and active states, shed lights on intersubunit interactions between GB1 and GB2 and ligand recognition, and propose models of Gi1 engagement to the receptor. We also observed multiple ordered cholesterols not only surrounding the periphery of both TM domains like many other GPCRs,38,39 but also between two subunits primarily in the inactive state (Supplementary information, Fig. S6a). Surprisingly, well-defined density with the shape of phospholipid in the TM bundle of each subunit was also observed in both states (Supplementary information, Fig. S6b–e). Considering the extensive interactions between each phospholipid and the corresponding subunit and the consistent presence of the phospholipids in both states, we speculated that phospholipids in the core pockets are more likely structural components.
Overall structure of GABAB receptor in the inactive state
The overall architecture of GABAB receptor reveals that heterodimeric GB1 and GB2 subunits face each other side by side with a pseudo C2-symmetry. The linker between extracellular VFT and TM domains within each subunit stands almost vertically to the bilayer. The extended ECL2 β-hairpin interacts extensively with the linker through three-stranded anti-parallel β-sheet, thus bridging VFT and TM domains in each subunit as a stalk (Figs. 1, 2a). This arrangement is similar to mGlu5 receptor,33 yet lacks the cysteine-rich domain leading to the shortened height of the receptor and partially contributing to the enhanced stability of the receptor in the inactive conformation initially observed by negative-stain EM (Supplementary information, Fig. S1d). The structure of the GABAB VFT domains in the context of full-length receptor is very similar to that of the corresponding crystal structures of soluble VFT in both inactive and active states as previously reported,29 with root mean squared deviation (r.m.s.d.) of 0.8 Å and 1.2 Å, respectively (Supplementary information, Fig. S7a, b).
a, b Orthogonal views of the superimposed structures of GABAB receptor in inactive and active states, showing the domain repositioning upon agonist binding-induced activation. Side views (a) and intracellular views (b) of superposed structures are shown, with the active structure in translucent in the left panels and the inactive structure in translucent in the right panels, respectively. VFT domains and loops are omitted for clarity in b. Red arrows indicate the translation direction and distance for GB1 and GB2 (measured at extracellular tips of TM1 helices), respectively. Structures were aligned on the combined domains of GB1 VFT and GB2 lobe 1, the relatively stable parts of the receptor along activation pathway.
CGP54626 anchors at the lobe 1 of GB1 VFT largely by hydrogen bonds and maintains the receptor in the inactive state (Supplementary information, Fig. S7d). Accordingly, both VFT modules in the structure of the antagonist-bound receptor are in an open state, resulting in the heterodimer interfaces of GABAB in the VFT region mainly through two lobe 1 with the buried surface area of 702 Å2 (Supplementary information, Fig. S8a). In addition, the interfaces in the inactive state also involve the TM domains. Backbone separation distance of GABAB between the TM5 helices are 8 Å in contrast to 17 Å of the inactive structure of mGlu5 (Supplementary information, Fig. S9a, b) and thus they make extensive contacts via lipid-mediated Van der Waals interactions at the extracellular half and salt bridging at intracellular half (Fig. 3a–c). Salt bridge network close to the cytoplasmic membrane surface is mediated by a charged residue quartet from the TM3 and TM5 helices, namely, H6893.55 and E7905.60 of GB1, H5793.55 and E6775.60 of GB2 (superscript refers to the GPCRdb numbering scheme).40 Moreover, we observed well-resolved and ordered densities along the TM interface and assigned them as putative cholesterols which connect a triplet of phenylalanine residues packing against one another in the proximal polar network (GB1a: F7865.56; GB2: F6705.53 and F6735.56) and a quintuplet of leucine residues in the middle of TM region (GB1a: L7795.49, L7825.52, L7835.53; GB2: L6665.49, L6695.52), all of which are from TM5 helices (Fig. 3b). At the extracellular half of the TM interface, TM5 helices facing each other through short uncharged side-chains form a deep crevice occupied by three putative cholesterols contributing to an additional buried surface area of 419 Å2, which further substantially stabilizes the receptor in the inactive conformation (Fig. 3a and Supplementary information, Fig. S6a). This novel TM interface represents the structural signature of GABAB heterodimer TM domains in the inactive conformation.
a, b Side (a) and intracellular (b) views of the inactive TM interface. EM densities (magenta) corresponding to putative cholesterols are shown in surface. c Close view of inactive TM interface proximal to the cytoplasm. H6893.55 and E7905.60 of GB1, H5793.55 and E6775.60 of GB2 forms salt bridge network to stabilize an inactive conformation. d Detailed interactions of the TM interface in the active state. e, f Cryo-EM density (e) and the detailed interactions (f) of (+)-BHFF (PAM) in the active TM interface. The critical residues are shown as sticks. Hydrogen bonds are depicted as dashed lines.
Conformational transition of GABAB heterodimer during activation
The 3.0 Å cryo-EM structure of the GABAB receptor in the active state reveals distinct structural configuration in both VFT and TM domains from that in the inactive state (Fig. 2a). Agonist baclofen anchors between two lobes of the GB1 VFT (Supplementary information, Fig. S7f) and induces the closure of the GB1 VFT, during which the lobe 2 of GB1 VFT undergoes a twist motion and contacts the opposite lobe 2 shortening the distance between the C-termini of the two VFT from 43 Å to 26 Å, in comparison to the distance changing from 45 Å to 32 Å as observed in the previous structural studies of the soluble VFT (Supplementary information, Fig. S7a, b). These conformational changes triggered by agonist binding propagate through the stalks and finally relay to the TM domains, leading to translations of the GB1 and GB2 TMs by 31 Å and 19 Å, respectively. Consequently, these structural transformations result in a TM6/TM6 interface (Figs. 2b, 3d), consistent with previous crosslinking studies.41 This interface is also observed in the agonist-bound structure of the mGlu5 homodimer,33 suggesting that the TM6/TM6 interface is likely a hallmark of the activation of the class C GPCR.
Akin to the TM interface in the inactive GABAB receptor, the interactions between the TM6/TM6 helices are strengthened by the hydrophobic and hydrophilic contacts. Compared to the lipid-mediated hydrophobic interactions at the extracellular half in the inactive state, the two TM6 helices in the active conformation directly pack together at their extracellular half via hydrophobic interactions, involving V8146.48, I8186.52, V8226.56, I8256.59, L826ECL3, A8327.27, and F8367.31 from GB1 and M6946.41, I7016.48, M7026.49, I7056.52, A7896.55, V7096.56, L7126.59, T713ECL3, Q716ECL3, and V7197.27 from GB2 and contributing to a buried surface area of 507 Å2 (Fig. 3d and Supplementary information, Fig. S8b). In the middle of the TM interface is a hydrogen bond network locked by the Y6.44 and N6.45 from both subunits (Fig. 3d and Supplementary information, Table S1). The TM6/TM6 interface is further stabilized by BHFF, a potent and selective GABAB receptor PAM agonist (PAM-ago), which can enhance the potency of endogenous agonist GABA and also directly activate GABAB receptor in the absence of other agonists with an unknown mechanism.42 The high-resolution cryo-EM map enables us to identify and unambiguously assign the BHFF into the density located in the cavity formed by the intracellular tips of TM5-TM6 of GB1 and TM6 of GB2 (Fig. 3e, f). Noteworthly, we found that the pure enantiomer (+)-BHFF fits into the density better than (−)-BHFF, in agreement with previous pharmacological studies suggesting that rac- and (+)-BHFF could enhance the GABA potency by 15.3-fold and 87.3-fold, respectively. The (+)-BHFF is mainly recognized by the hydrophobic interactions, involving A7885.58, Y7895.59, M8076.41, and Y8106.44 of GB1 and K6906.37, Y6916.38, and M6946.41 of GB2. Additionally, 3-hydroxy group and ketone are hydrogen-bonded to K792ICL3 of GB1 (Fig. 3f). We thus identified a novel PAM-binding pocket within a GPCR dimer and provide a template for future structure-based drug design of allosteric modulators targeting the dimeric interface of class C GPCR.
Structure and activation of TM domain of GABAB receptor
All the available TM structures of class C GPCRs are in the apo or inactive conformation, including cryo-EM structure of the full-length mGlu5 bound to agonist.33 Superposition of individual TM domains between GABAB and other class C GPCRs including mGlu1 and mGlu5 reveals distinct structural features for TM5, TM7, ECL1, ECL2 and ICL3 (Supplementary information, Fig. S9c). Extracellular tips of TM5 and TM7 helices of GABAB receptor demonstrate outward shifting by 5 Å, likely resulting from the presence of endogenous phospholipid in the center of TM bundle which extensively contacts TM3, 5, 6 and 7 (Supplementary information, Fig. S6a). The ECL2 of GABAB spanning a β-hairpin configuration is extended by 5-residue relative to other class C GPCRs, leading to more tight interaction with the linker and enabling the direct contacts with the bottom of VFT (Fig. 4 and Supplementary information, Fig. S9c). Therefore, this extension facilitates the agonist binding-induced conformational changes of VFT conducting to TM domain. Moving to the intracellular loops, the most profound difference is the position of ICL3, which is proposed to play a crucial role in G-protein coupling.25 The ICL3 of GABAB receptor is comprised of 11 residues for both subunits (residues 791–801 for GB1 and residues 678–688 for GB2) and stretched over the TM pocket at the cytoplasmic surface. The conformation of ICL3 is stabilized by the polar network formed by S51512.53, N5202.39, K5743.50, N6876.34 and D6886.35, which are highly conserved among class C GPCRs, and further strengthened by hydrophobic interactions between L51112.49, M51412.52, A6755.58, W6765.59 (Supplementary information, Fig. S10a–c). We anticipate that the conformation of ICL3 in GABAB receptor restricts the access of G proteins or β-arrestins to the receptor, similarly to helix 8 of the angiotensin II receptor AT2R.43 In contrast, the same loop from mGlu1 and mGlu5 is 2-residue shorter and stands almost perpendicular to the bilayer, apparently leaving the TM cavity at the cytoplasmic side more accessible to the effector proteins (Supplementary information, Fig. S9c).
The overall structures of the GABAB TM domains are highly similar in the inactive state with r.m.s.d. of 0.94 Å as expected, considering that the two TM domains share 72% sequence homology. Although upon activation the apical tip of GB1 ECL2 shifts 6 Å measured at Cα atom of I750ECL2, the extracellular conformational changes induced by agonist binding do not relay to GB1 TM domain through the stalk, which is mostly unchanged (r.m.s.d. of 2.23 Å) (Fig. 4a). In contrast, substantial conformational changes occur at the GB2 stalk region (r.m.s.d. of 3.94 Å), translating by 5.6 Å measured at I469, which triggers the extracellular tips of TM4 and TM5 shifting 2.0 Å and 2.9 Å to the opposite direction measured at D6194.54 and T6545.37, respectively (Fig. 4b). Consequently, the intracellular tip of GB2 TM3 undergoes 5.2 Å shifting (measured at Cα atom of I5813.57) towards TM4 and TM2, leading to a 2.7 Å and 1.7 Å outward movement measured at Cα atoms of L5984.33 and M5192.38, respectively. The most profound conformational changes at the cytoplasmic half locate at TM5-ICL3. GB2 TM5 extends one helical turn and shifts 5.4 Å measured at Cα atom of W6785.61 toward TM3, resulting in the breaking of the ionic interactions among N6876.34, D6886.35and S51512.53, N5202.39, K5743.50, and thus the ICL3 stands up, uncovering the lid of the TM pocket at the cytoplasmic side (Supplementary information, Fig. S10b). Taken together, the orchestration of the conformational changes in the TM domain of GB2 subunit enables it to accommodate the downstream Gi/o protein to initiate the intracellular signaling.
Gi heterogeneous coupling to GABAB heterodimer
We identified three thermostable conformations demonstrating evident density for Gi1 protein, which are B2a, B2b and B1 states with the population distribution of 74%, 9% and 17%, respectively, among the sub-dataset (Fig. 5a–c; Supplementary information, Fig. S3). This observation suggests that Gi1 protein predominantly couples to GB2 subunit, consistent with the asymmetric activation model of GABAB receptor.21 It is noteworthy that the density corresponding to the α-helical domain (AHD) of the Gαi in each state is separated from the Gαi Ras-like domain, which was also observed in many other activated GPCR–Gi complexes,44,45,46,47 suggesting that Gi1 is likely in its activated and nucleotide-free conformation in three observed conformations. The structural rearrangement of GB2 TM domain barely exposes residues from TM helices and thus creates a shallow binding site for the heterotrimeric Gi1 protein (Fig. 5a–d), whereas all the available GPCR–G structures show a deep cavity formed by the outward movement of the cytoplasmic half of TM6 together with TM2, 3, 5, and 7.48 This remarkable shallow pocket disallows the insertion of the α5-helix of the Gαi Ras-like domain into the receptor TM domain and is probably responsible for the conformational heterogeneity present in the GABAB–Gi1 complex. Superposing the isolated GB1–Gi structure from B1 state to the heterodimer structure of either B2a or B2b state shows the potential steric hindrance between the two Gi1 proteins, perfectly explaining why only one G protein binds to the GABAB receptor at a time (Fig. 5e).
a–c Cryo-EM density maps and the models of the GABAB–Gi1 complex in B2a state (a), B2b state (b), and B1 state (c). GB1 in red, GB2 in green, Gαi in gold, Gβ in cyan, Gγ in purple and scFv16 in gray. d Magnified view of the interaction between the α5-helix of the Gαi and GABAB receptor in B2a state is shown. Intracellular loops 1, 2, and 3 are highlighted. e Superposition of the B1 state with either B2a state or B2b state showing the potential steric clash between Gi proteins, indicating that only one Gi protein can bind to GABAB receptor at one time. Gi in B2a state, brown; Gi in B2b state, blue; Gi in B1 state, gray.
The interface between the activated GABAB receptor and Gi1 protein in B2a state involves the ICLs and the intracellular tip of the TM3 of the receptor and the α5-helix of the Gαi Ras-like domain (Fig. 5d). The carboxyl terminal α5-helix of the Gαi overlaps the position of GB2 ICL3 in the inactive state, explaining the requirement for GB2 ICL3 opening to accommodate the Gi/o protein. Previous studies showed that the single mutation L686P in the ICL3 of GB2 suppresses the activation of G protein in either HEK293 cells or cultured neurons, highlighting the critical role of ICL3 of GB2 for the coupling of the heterodimeric GABAB receptor to G-proteins,25 in agreement with our structural findings. Interestingly, the α5-helix of the Gαi in B2a state translates downwards by ~8 Å relative to that of class A GPCR–Gi complex in both canonical and noncanonical states (Supplementary information, Fig. S10d), leading to the absence of Gβγ in the receptor–Gi1 interface. Additionally, Gi1 in B2b and B1 states is rotated by about ~90° to the opposite side of that in B2a state relative to the receptor, positioning Gi1 in the complex in distinct arrangements (Supplementary information, Fig. S10e). Surprisingly, unlike class A, B and F GPCRs, TM6 holds its position in response to activation, constrained by the TM6/TM6 interface in the active state (Supplementary information, Fig. S10f).
The observation of the GABAB–Gi1 complex in GB1 state provides the structural explanation of the basal activity of GABAB receptor given that GB1 TM domain shares the same conformation in the inactive and active states, in agreement with the observation that expression of GB1 alone at the cell surface activates ERK1/2 through Gi/o-dependent pathway.27,28 Considering the significant difference in population between the two B2 states, we anticipate that B2a is likely more thermodynamically stable and therefore forms after the B2b state along the activation pathway of G protein. A similar phenomenon was observed in the neurotensin receptor 1(NTSR1)–Gi1 complex.49
Discussion
GABAB receptor represents the first example of an obligatory GPCR heterodimer; it functions as an allosteric multi-domain protein and oscillates between the inactive and active states.21 Ligands or G proteins interplay with the pre-existing states of the receptor based on the “selection mode of allostery”.21,50 In this study, by synchronizing receptor in the inactive state in the presence of antagonist or completely stabilizing receptor in the active state in the presence of agonist, PAM and Gi1 protein, we determined the atomic-resolution cryo-EM structures of a full-length human GABAB heterodimer in both inactive and active states. The high-resolution cryo-EM maps not only confirmed the binding mode of CGP54626 and baclofen observed in the crystal structures of GABAB VFT,29 but also revealed the working mechanism of BHFF as PAM agonist, which stabilizes a novel intersubunit interface in the TM domains of the active GABAB receptor. Surprisingly, endogenous phospholipids settle inside each TM extracellular pore, equivalent to the classical orthosteric binding pocket for class A GPCR, and lead to a distinct conformational configuration of GABAB TM domains compared to other class C GPCRs, suggesting that these lipids may function as structural components, which is probably a unique phenomenon to GABAB receptor.
Structural comparison of GABAB receptor in different states enabled us to propose a structural rearrangement for GABAB receptor activation. Agonist binding induces the compaction of two VFTs (Fig. 2; Supplementary information, Fig. 7a, b), propagating through the stalk domains to the TM domains, which in turn reorients the TM interface between two subunits from TM3-TM5/TM3-TM5 in the inactive state to TM6/TM6 in the active state, in perfect agreement with previous crosslinking data for GABAB receptor and mGlu homodimers.41,51 Therefore, the overall domain rearrangements upon activation of GABAB receptor resemble the published cryo-EM structures of mGlu5,33 suggesting that this reorientation of TM domains is possibly the hallmark of class C GPCR activation.
We determined cryo-EM structures of a GPCR heterodimer in a complex with Gi1 protein, which only can couple to either of the subunit at one time to avoid steric hindrance by the presence of the two Gi1 proteins. The unprecedented structures reveal novel conformational changes within the GB2 TM domain to accommodate the Gi protein involving the opening of ICL3 and movement of TM3, 4 and 5 helices, enabling GABAB receptor to predominantly couple to Gi1 protein through GB2 subunit. However, the structure of GABAB–Gi1 complex coupling via GB1 demonstrates that GB1 couples to Gi protein at certain circumstance. Indeed, GB1 in Caenorhabditis elegans directly couples to Gi/o protein-dependent signaling,52,53 indicating that GB1 retains the capacity for Gi protein coupling during evolution. Common activation mechanism for G protein activation by GPCRs has been proposed, in which the landmark of GPCR activation is the outwards movement of TM6 forming a cavity to accommodate the G proteins or β-arrestins, reflecting the convergence of activation pathways in class A, B and F GPCRs.48,54,55 However, the GABAB–Gi1 engagement patterns have not been observed elsewhere, implying the complex of G protein coupling to GPCRs, especially for GPCRs in dimeric form. Further studies are required to determine the high-resolution structure of GABAB–Gi1 complex to decipher the molecular mechanism of GABAB–G protein assembly. Additional structural studies are also needed to determine whether similar G protein engagement is present in other class C GPCRs.
Materials and methods
Constructs
Human GABAB receptor with the HA signal peptide including GB1a (UniProt: Q9UBS5) and GB2 (UniProt: O75899) were cloned into pEG BacMam vector.56 To facilitate expression and purification, an 8× histidine tag and 3C protease cleavage site were inserted at the C-terminus of GB1a subunit, while a Flag epitope tag (DYKDDDD) and a 2× GSG linker were added to the N-terminus of GB2 subunit. We utilized different constructs with cleavable C-terminal domains: GB1 and GB2 were truncated after coiled-coil domain; GB1Δcc and GB2Δcc were truncated before coiled-coil domain; GB1ΔC means that the C-terminus of GB1 was removed. In addition, we constructed the Dual pEG BacMam vector, which could express GB1 and GB2 together.
Expression and purification of inactive GABAB receptor
GB1a (GB1cc residues 15–919) and GB2 (GB2cc residues 42–819) were co-expressed in HEK293F cells. In brief, purified plasmid DNA was mixed with PEI 25K at a 3:1 ratio of PEI to DNA (w/w) followed by addition to HEK293F cells when the density reached ~2.8 million/mL.57 10 mM sodium butyrate was added after 16–18 h post-infection, and cells were then grown for 3 days at 30 °C before harvest. The infected cells were collected by centrifugation at 1000× g for 15 min and washed once with 1× PBS buffer. Cells were suspended in 50 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol, 2 mM MgCl2, 20 μM CGP54626 (Tocris Bioscience) with protease inhibitor cocktail followed by homogenization. The membrane was solubilized for 3 h at 4 °C with 0.5% (w/v) lauryl maltoseneopentyl glycol (LMNG, Anatrace), 0.1% (w/v) cholesteryl hemisuccinate (CHS, Anatrace). After centrifugation at 30,000× g for 30 min, the supernatant was bound to Ni-NTA column and further loaded onto M1 anti-FLAG affinity resin. The protein was eluted in elution buffer consisting of 50 mM HEPES, pH 7.5, 150 mM NaCl, 0.01% LMNG, 0.002% CHS, 20 μM CGP54626, 5 mM EGTA and 0.1 mg/mL FLAG peptide. The GABAB receptor was concentrated in a 100-kDa cutoff Vivaspin (Millipore) filter and run on a Superose™ 6 Increase column (GE Healthcare).
Expression of heterotrimeric Gi
Heterotrimeric Gi was expressed as previously described.58 In general, the dominant-negative Gαi1 was introduced four mutations (S47N, G203A, E245A and A326S) and a 6× histidine tag was added at the N-terminus of the β subunit. The Sf9 insect cells (Expression Systems) at a density of 2.4 million/mL were infected with both Gαi and Gβγ virus at a 1:1 ratio. Cells were harvested 48 h after infection and collected by centrifugation at 1000× g for 15 min. Finally, the cells were washed once with 1× PBS buffer and snap frozen in liquid nitrogen for later use.
Expression and purification of scFv16
The secreted scFv16 was expressed and purified by using the bac-to-bac system as previously described.36 In brief, the virus of scFv16 with 6× histidine tag at the C-terminus was infected in Trichoplusia ni Hi5 insect cells for 48 h. The pH of the supernatant was balanced by the addition of Tris pH 8.0 while chelating agents were removed by addition of 1 mM nickel chloride and 5 mM calcium chloride. After incubation with stirring at room temperature for 1 h, the supernatant was loaded onto Ni-NTA resin and further eluted in elution buffer consisting of 20 mM HEPES, pH 7.5, 500 mM NaCl, and 250 mM imidazole. The sample was first treated with 3C protease, and then diluted and reloaded onto the Ni-NTA column to remove the 6× histidine tag. The flow-through was collected and purified by gel filtration chromatography using a Superdex 200 column. Finally, the concentrated of scFv16 was flash frozen in liquid nitrogen until further use.
Formation and purification of the GABAB-Gi-scFv16 complex
GB1a (GB1ΔC residues 15–860) and GB2 (GB2Δcc residues 42–780) were chosen to form GABAB-Gi1 complex. The conditions for GABAB expression in HEK293F cells were described earlier. Purified plasmid DNA mixed with PEI 25K at a 3:1 ratio of PEI to DNA (w/w) was added to HEK293F cells when the density reached ~2.8 million/mL. The infected HEK293F cells and the expressed Gi1 cells together were suspended and disrupted in 50 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol, 2 mM MgCl2, 100 μM baclofen and 50 μM BHFF (Tocris Bioscience) with protease inhibitor cocktail followed by addition of 50 mU/mL apyrase and 1.0 mg scFv16. The membrane was solubilized with 0.5% (w/v) LMNG and 0.1% (w/v) CHS for 3 h at 4 °C and separated by centrifugation at 30,000× g for 30 min. The supernatant was bound to Ni-NTA column and further loaded onto M1 anti-FLAG affinity resin. The protein was eluted in elution buffer consisting of 50 mM HEPES, pH 7.5, 150 mM NaCl, 0.01% LMNG, 0.002% CHS, 100 μM baclofen and 50 μM BHFF, 5 mM EGTA and 0.1 mg/mL FLAG peptide. The GABAB-Gi-SCFV16 complex was concentrated in a 100-kDa cutoff Vivaspin (Millipore) filter and run on a Superose™ 6 Increase column.
G protein pull-down analysis
Three different group truncations of GABAB receptor were cloned into the pEG BacMam vector, respectively. Flag epitope tag (DYKDDDD) and a 2× GSG linker were added to the N-terminus of GB2. Different group truncations of GABAB receptor were expressed in 30 mL HEK293F cells while heterotrimeric Gi was expressed in 30 mL Sf9 insect cells. These cells were suspended in 50 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol, 2 mM MgCl2, 100 μM baclofen and 50 μM BHFF with protease inhibitor cocktail followed by homogenization. 50 mU/mL apyrase and 0.03 mg scFv16 were added to incubate for 3 h. The membrane was solubilized with 0.5% (w/v) LMNG and 0.1% (w/v) CHS for another 3 h at 4 °C. After centrifugation, the supernatant was incubated with 25 μL M1 anti-FLAG affinity resin for 1 h at 4 °C and then eluted in 50 μL elution buffer supplemented with 100 μM baclofen and 50 μM rac-BHFF, 5 mM EGTA and 0.1 mg/mL FLAG peptide. Samples collected from different truncations were analyzed by SDS-PAGE.
Bioluminescence resonance energy transfer (BRET) assay
HEK293 cells were transfected with wild-type or truncation (GB1ΔC-GB2Δcc) GABAB receptor, Gβ1, Venus-Gγ2 and Gαi-Rluc8 by Lipofectamine 2000 and split into 96-well flat-bottomed white microplates. After 24 h transfection, cells were washed and starved in PBS at 37 °C for 1 h. BRET measurements were performed using the Mithras LB 940 (Berthold Technologies, German). The signals emitted by the donor (460–500 nm band-pass filter, Em 480) and the acceptor entity (510–550 nm band-pass filter, Em 530) were recorded after the addition of 5 μM Coelenterazine H. All measurements were performed at 37 °C. The BRET signal was determined by calculating the ratio of the Em 530 and Em 480. The net BRET ratio was defined as the experimental BRET signal values with the baseline subtracted (basal BRET ratio), which was recorded before the stimulation of cells. For dose–response experiments, data were analyzed using nonlinear curve fitting for the log (agonist) vs. Response (three parameters) curves in GraphPad Prism software.
Cryo-EM grid preparation and data collection
For the preparation of Cryo-EM grids, 3 μL of the purified antagonist-bound GABAB heterodimer or baclofen/BHFF-bound GABAB–Gi1 complex at ~2.0 mg/mL was applied onto a glow-discharged 200 mesh holey carbon grid (Quantifoil R1.2/1.3). The grids were blotted and then plunge-frozen in liquid ethane using Vitrobot Mark IV (Thermo Fischer Scientific). Cryo-EM data collection was performed on a Titan Krios at 300 kV accelerating voltage in the Center of Cryo-Electron Microscopy, Zhejiang University (Hangzhou, China). Micrographs were recorded using a Gatan K2 Summit direct electron detector in counting mode with a nominal magnification of 29,000×, corresponding to a pixel size of 1.014 Å. Image stacks was obtained at a dose rate of ~8.0 electrons per Å2 per second with a defocus ranging from −0.5 to −2.5 μm. The total exposure time was 8 s and intermediate frames were recorded in 0.2 s intervals, resulting in an accumulated dose of 64 electrons per Å2 and a total of 40 frames per micrograph. A total of 4740 and 4624 movies were collected for the antagonist-bound GABAB heterodimer and baclofen/BHFF-bound GABAB–Gi1 complex, respectively.
Imaging processing and 3D reconstruction
For the dataset of antagonist-bound GABAB heterodimer, image stacks were subjected to beam-induced motion correction using MotionCor2.1.59 Contrast transfer function (CTF) parameters for each non-dose weighted micrograph were determined by Gctf.60 Semi-automated particle selection was performed using RELION-3.0-beta2,61 yielding 2,969,413 particles. The particles were extracted on a binned dataset with a pixel size of 2.028 Å and imported to CryoSPARC v2.4262 for 2 rounds of 2D classification. The poorly defined 2D classes were discarded, producing 1,757,222 particles for further abinitio reconstruction and heterogeneous refinement. After 2 rounds of heterogeneous refinement, the well-defined subset with 802,381 particles were re-extracted with a pixel size of 1.014 Å in Relion. The particles were subsequently subjected to 3D classification, which produced two good subsets accounting for 610,689 particles. Further 3D classifications focusing the alignment on the complex and the TMD domains, produced high-quality subsets, accounting for 374,595 particles, which were subsequently subjected to 3D refinement and Bayesian polishing. The overall refinement generated a map with an indicated global resolution of 3.0 Å at a Fourier shell correlation of 0.143. To further improve the map quality, local refinement focusing on the TMD and VFT domains were performed in Relion. The locally refined map for the VFT and TMD show a global resolution of 2.8 Å and 3.0 Å, which were merged using vop maximum command in UCSF Chimera.63 This composite map was then “Z-flip” to get the correct handedness and used for subsequent model building and analysis.
For the dataset of GABAB-Gi1 complex, movies were subjected to beam-induced motion correction using MotionCor2.159 and CTF estimation using Gctf.60 Template-based automated particle selection was performed using RELION-3.0-beta2, producing 3,075,533 particles for further 2D classification in CryoSPARC. The well-defined classes with 1,471,649 particles were selected for abinitio reconstruction and heterogeneous refinement, resulting in 488,801 particles with good receptor density. The subset was further re-extracted on an unbinned dataset in Relion and subjected to multiple rounds of 3D classification. Three classes showing different conformations of G protein binding (B1, B2a and B2b) were identified, accounting for 30,542 particles, 130,901 particles and 16,048 particles, respectively. In contrast, the other three classes showed the fuzzy density of the G protein. The subsets for the 3 conformational states were further subjected to 3D refinement and Bayesian polishing, yielding density maps with indicated global resolutions of 8.8 Å, 6.8 Å, and 8.6 Å, respectively. To find out the TMD conformations of the G protein-bound GABAB, the map of the dominant conformation, B2a, was further refined with a mask on the receptor. The locally refined density map shows a relatively clear density for the TM bundle arrangement with an indicated resolution of 5.8 Å. Due to the high consistency of the receptor conformations in all 3D classes including three identified G protein-bound states and three poorly defined G protein-bound classes, we reasoned that the receptor adopts the same or highly similar conformation regardless of the orientation of G protein engagement. We then focused the alignment on the receptor and performed 3D classification with a mask on the receptor and TMD, respectively. The subsets, accounting for 237,606 particles, showing the high-quality EM density were selected and were subjected to 3D refinement and Bayesian polishing. The refinement generated a map with an indicated global resolution of 3.1 Å. To improve the quality of TMD domain, local refinement focusing on the TMD and VFT domains was further performed in Relion. The locally refined map for the VFT and TMD show a global resolution of 2.8 Å and 3.2 Å, which were merged in UCSF Chimera and used for subsequent model building and analysis. Local resolution was determined using the Bsoft package64 with half maps as input maps.
Model building and refinement
The initial template of the TMDs in GB1 and GB2 was generated using SWISS-MODEL.65 The initial template of VFT domain was derived from the crystal structure of antagonist-bound GABAB VFT (PDB: 4MR7).29 Antagonist, agonist and PAM coordinates and geometry restraints were generated using phenix.elbow.66 Models of TMD and VFT domains were docked into the EM density map of antagonist-bound GABAB using UCSF Chimera.63 The initial model was subjected to iterative manual rebuilding in COOT67 and real-space refinement in PHENIX.66 The final refinement statistics were validated using the module “comprehensive validation (cryo-EM)” in PHENIX. The refined model of antagonist-bound GABAB was used as the initial model for baclofen/BHFF-bound GABAB. Structures of TMD and VFT lobe domains of GB1 and GB2 were docked into the density map using UCSF Chimera, respectively. The model was further subjected to manual rebuilding and real-space refinement. The structures of baclofen/BHFF-bound GABAB and Gil protein from the structure of human cannabinoid receptor 2-Gi1 complex (PDB code: 6PT0)44 were used to generate the docked model of GABAB-Gi complex in B1, B2a and B2b states, respectively. The refinement statistics of the inactive and active GABAB heterodimer are provided in Supplementary information, Table S2. Structure figures were created using UCSF Chimera,63 and the UCSF Chimera X package.68
Data availability
Cryo-EM maps have been deposited in the Electron Microscopy Data Bank under accession codes: EMD-30301 (CGP54626-bound GABAB receptor), EMD-30300 (baclofen/BHFF-bound GABAB receptor in the presence of Gi1 protein). The atomic coordinates have been deposited in the Protein Data Bank under accession codes: 7C7S (CGP54626-bound GABAB receptor) and 7C7Q (baclofen/BHFF-bound GABAB receptor in the presence of Gi1 protein).
References
Barnard, E. A. et al. International Union of Pharmacology. XV. Subtypes of gamma-aminobutyric acidA receptors: classification on the basis of subunit structure and receptor function. Pharmacol. Rev. 50, 291–313 (1998).
Bettler, B. & Tiao, J. Y. Molecular diversity, trafficking and subcellular localization of GABAB receptors. Pharmacol. Ther. 110, 533–543 (2006).
Kaupmann, K. et al. GABA(B)-receptor subtypes assemble into functional heteromeric complexes. Nature 396, 683–687 (1998).
Mott, D. D. & Lewis, D. V. The pharmacology and function of central GABAB receptors. Int. Rev. Neurobiol. 36, 97–223 (1994).
Bettler, B., Kaupmann, K., Mosbacher, J. & Gassmann, M. Molecular structure and physiological functions of GABA(B) receptors. Physiol. Rev. 84, 835–867 (2004).
Luscher, C., Jan, L. Y., Stoffel, M., Malenka, R. C. & Nicoll, R. A. G protein-coupled inwardly rectifying K+ channels (GIRKs) mediate postsynaptic but not presynaptic transmitter actions in hippocampal neurons. Neuron 19, 687–695 (1997).
Tu, H. et al. GABAB receptor activation protects neurons from apoptosis via IGF-1 receptor transactivation. J. Neurosci. 30, 749–759 (2010).
Lin, X. et al. An activity-based probe reveals dynamic protein-protein interactions mediating IGF-1R transactivation by the GABA(B) receptor. Biochem. J. 443, 627–634 (2012).
Samanta, D. & Zarate, Y. A. Widening phenotypic spectrum of GABBR2 mutation. Acta Neurol. Belg. 119, 493–496 (2019).
Dalmau, J. & Graus, F. Antibody-mediated encephalitis. N. Engl. J. Med. 378, 840–851 (2018).
Hoftberger, R. et al. Encephalitis and GABAB receptor antibodies: novel findings in a new case series of 20 patients. Neurology 81, 1500–1506 (2013).
Froestl, W. Chemistry and pharmacology of GABAB receptor ligands. Adv. Pharmacol. 58, 19–62 (2010).
Addolorato, G. et al. Baclofen efficacy in reducing alcohol craving and intake: a preliminary double-blind randomized controlled study. Alcohol Alcohol. 37, 504–508 (2002).
Chang, E. et al. A review of spasticity treatments: pharmacological and interventional approaches. Crit. Rev. Phys. Rehabil. Med. 25, 11–22 (2013).
Xu, X. M., Wei, Y. D., Liu, Y. & Li, Z. X. Gamma-hydroxybutyrate (GHB) for narcolepsy in adults: an updated systematic review and meta-analysis. Sleep Med. 64, 62–70 (2019).
Kniazeff, J., Prezeau, L., Rondard, P., Pin, J. P. & Goudet, C. Dimers and beyond: the functional puzzles of class C GPCRs. Pharmacol. Ther. 130, 9–25 (2011).
Romano, C., Yang, W. L. & O’Malley, K. L. Metabotropic glutamate receptor 5 is a disulfide-linked dimer. J. Biol. Chem. 271, 28612–28616 (1996).
Bai, M., Trivedi, S. & Brown, E. M. Dimerization of the extracellular calcium-sensing receptor (CaR) on the cell surface of CaR-transfected HEK293 cells. J. Biol. Chem. 273, 23605–23610 (1998).
White, J. H. et al. Heterodimerization is required for the formation of a functional GABA(B) receptor. Nature 396, 679–682 (1998).
Kuner, R. et al. Role of heteromer formation in GABAB receptor function. Science 283, 74–77 (1999).
Pin, J. P. & Bettler, B. Organization and functions of mGlu and GABAB receptor complexes. Nature 540, 60–68 (2016).
Pagano, A. et al. C-terminal interaction is essential for surface trafficking but not for heteromeric assembly of GABA(b) receptors. J. Neurosci. 21, 1189–1202 (2001).
Ng, G. Y. et al. Identification of a GABAB receptor subunit, gb2, required for functional GABAB receptor activity. J. Biol. Chem. 274, 7607–7610 (1999).
Malitschek, B. et al. The N-terminal domain of gamma-aminobutyric Acid(B) receptors is sufficient to specify agonist and antagonist binding. Mol. Pharm. 56, 448–454 (1999).
Duthey, B. et al. A single subunit (GB2) is required for G-protein activation by the heterodimeric GABA(B) receptor. J. Biol. Chem. 277, 3236–3241 (2002).
Robbins, M. J. et al. GABA(B2) is essential for g-protein coupling of the GABA(B) receptor heterodimer. J. Neurosci. 21, 8043–8052 (2001).
Richer, M. et al. GABA-B(1) receptors are coupled to the ERK1/2 MAP kinase pathway in the absence of GABA-B(2) subunits. J. Mol. Neurosci. 38, 67–79 (2009).
Baloucoune, G. A. et al. GABAB receptor subunit GB1 at the cell surface independently activates ERK1/2 through IGF-1R transactivation. PLoS ONE 7, e39698 (2012).
Geng, Y., Bush, M., Mosyak, L., Wang, F. & Fan, Q. R. Structural mechanism of ligand activation in human GABA(B) receptor. Nature 504, 254–259 (2013).
Muto, T., Tsuchiya, D., Morikawa, K. & Jingami, H. Structures of the extracellular regions of the group II/III metabotropic glutamate receptors. Proc. Natl. Acad. Sci. USA 104, 3759–3764 (2007).
Wu, H. et al. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science 344, 58–64 (2014).
Dore, A. S. et al. Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature 511, 557–562 (2014).
Koehl, A. et al. Structural insights into the activation of metabotropic glutamate receptors. Nature 566, 79–84 (2019).
Jones, K. A. et al. GABA(B) receptors function as a heteromeric assembly of the subunits GABA(B)R1 and GABA(B)R2. Nature 396, 674–679 (1998).
Malherbe, P. et al. Characterization of (R,S)-5,7-di-tert-butyl-3-hydroxy-3-trifluoromethyl-3H-benzofuran-2-one as a positive allosteric modulator of GABAB receptors. Br. J. Pharmacol. 154, 797–811 (2008).
Koehl, A. et al. Structure of the micro-opioid receptor-Gi protein complex. Nature 558, 547–552 (2018).
Margeta-Mitrovic, M., Jan, Y. N. & Jan, L. Y. Function of GB1 and GB2 subunits in G protein coupling of GABA(B) receptors. Proc. Natl. Acad. Sci. USA 98, 14649–14654 (2001).
Zhao, L. H. et al. Structure and dynamics of the active human parathyroid hormone receptor-1. Science 364, 148–153 (2019).
Ma, S. et al. Molecular basis for hormone recognition and activation of corticotropin-releasing factor receptors. Mol. Cell 77, 669–680 (2020).
van der Kant, R. & Vriend, G. Alpha-bulges in G protein-coupled receptors. Int. J. Mol. Sci. 15, 7841–7864 (2014).
Xue, L. et al. Rearrangement of the transmembrane domain interfaces associated with the activation of a GPCR hetero-oligomer. Nat. Commun. 10, 2765 (2019).
Koek, W., Cheng, K. & Rice, K. C. Discriminative stimulus effects of the GABAB receptor-positive modulator rac-BHFF: comparison with GABAB receptor agonists and drugs of abuse. J. Pharmacol. Exp. Ther. 344, 553–560 (2013).
Zhang, H. et al. Structural basis for selectivity and diversity in angiotensin II receptors. Nature 544, 327–332 (2017).
Xing, C. et al. Cryo-EM structure of the human cannabinoid receptor CB2-G(i) signaling complex. Cell 180, 645–654 (2020).
Koehl, A. et al. Structure of the µ-opioid receptor-G(i) protein complex. Nature 558, 547–552 (2018).
Kato, H. E. et al. Conformational transitions of a neurotensin receptor 1-G(i1) complex. Nature 572, 80–85 (2019).
Kang, Y. et al. Cryo-EM structure of human rhodopsin bound to an inhibitory G protein. Nature 558, 553–558 (2018).
Glukhova, A. et al. Rules of engagement: GPCRs and G proteins. ACS Pharmacol. Transl. Sci. 1, 73–83 (2018).
Kato, H. E. et al. Conformational transitions of a neurotensin receptor 1-Gi1 complex. Nature 572, 80–85 (2019).
Xu, C., Zhang, W., Rondard, P., Pin, J. P. & Liu, J. Complex GABAB receptor complexes: how to generate multiple functionally distinct units from a single receptor. Front. Pharmacol. 5, 12 (2014).
Xue, L. et al. Major ligand-induced rearrangement of the heptahelical domain interface in a GPCR dimer. Nat. Chem. Biol. 11, 134–140 (2015).
Yuan, F. et al. GABA receptors differentially regulate life span and health span in C. elegans through distinct downstream mechanisms. Am. J. Physiol. Cell Physiol. 317, C953–C963 (2019).
Chun, L. et al. Metabotropic GABA signalling modulates longevity in C. elegans. Nat. Commun. 6, 8828 (2015).
Qi, X. et al. Cryo-EM structure of oxysterol-bound human Smoothened coupled to a heterotrimeric Gi. Nature 571, 279–283 (2019).
Krumm, B. & Roth, B. L. A structural understanding of class B GPCR selectivity and activation revealed. Structure 28, 277–279 (2020).
Goehring, A. et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585 (2014).
Subedi, G. P., Johnson, R. W., Moniz, H. A., Moremen, K. W. & Barb, A. High yield expression of recombinant human proteins with the transient transfection of HEK293 cells in suspension. J. Vis. Exp. 106, e53568 (2015).
Liu, P. et al. The structural basis of the dominant negative phenotype of the Gαi1β1γ2 G203A/A326S heterotrimer. Acta Pharmacol. Sin. 37, 1259–1272 (2016).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H. Processing of structurally heterogeneous cryo-EM data in RELION. Methods Enzymol. 579, 125–157 (2016).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Heymann, J. B. Single particle reconstruction and validation using Bsoft for the map challenge. J. Struct. Biol. 204, 90–95 (2018).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Acknowledgements
We are grateful to H. Eric Xu for generously providing the plasmids of the heterotrimeric G protein. We thank S. Chang for technical support in cryo-EM data collection at the Center of Cryo-Electron Microscopy, Zhejiang University; Protein expression and purification were performed at the Protein Facilities, Zhejiang University School of Medicine with the support of C. Ma. J.L. was supported by the Ministry of Science and Technology (2018YFA0507003), the National Natural Science Foundation of China (81720108031, 81872945, 31721002 and 31420103909), the Program for Introducing Talents of Discipline to the Universities of the Ministry of Education (B08029), and the Mérieux Research Grants Program of the Institut Mérieux. Y.Z. was supported by the National Natural Science Foundation of China (81922071), the National Key Basic Research Program of China (2019YFA0508800), Zhejiang Province Natural Science Fund for Excellent Young Scholars (LR19H310001) and the Fundamental Research Funds for the Central Universities (2019XZZX001-01-06).
Author information
Authors and Affiliations
Contributions
C.S. designed the constructs, expressed and purified the antagonist-bound GABAB and the agonist/PAM-stimulated GABAB–Gi1 complex for cryo-EM data collection with the help of S.Z.; C.S., R.Z. and C.L. performed pull-down assay; C.L. and L.-N.C. expressed and purified scFv16; C.X. performed BRET assay; D.-D.S. evaluated the sample by negative-stain EM; C.M. prepared the cryo-EM grids; C.M., D.-D.S. collected the cryo-EM data; C.M. performed cryo-EM map calculation, model building and structure refinement; C.M. and C.S. analyzed the structures and prepared the figures with the help of D.-D.S., Q.S. and Z.J.; Y.Z. and J.L. conceived and supervised the project, analyzed the structures, and wrote the manuscript with inputs from all the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Mao, C., Shen, C., Li, C. et al. Cryo-EM structures of inactive and active GABAB receptor. Cell Res 30, 564–573 (2020). https://doi.org/10.1038/s41422-020-0350-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41422-020-0350-5
This article is cited by
-
Specific pharmacological and Gi/o protein responses of some native GPCRs in neurons
Nature Communications (2024)
-
G protein-coupled receptors (GPCRs): advances in structures, mechanisms, and drug discovery
Signal Transduction and Targeted Therapy (2024)
-
Constitutive activation mechanism of a class C GPCR
Nature Structural & Molecular Biology (2024)
-
Mechanism of sensitivity modulation in the calcium-sensing receptor via electrostatic tuning
Nature Communications (2022)
-
G protein coupling and activation of the metabotropic GABAB heterodimer
Nature Communications (2022)