Abstract
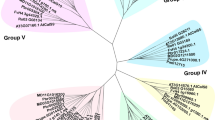
Cullin (CUL) gene family organizes the largest class of RING E3 ligases which play crucial roles in the ubiquitin-proteasome system to affect plant physiological activities. However, little is known about the CUL genes in fruit trees. In this study, 57 CUL genes were isolated from the sequenced genomes of seven Rosaceae trees and were divided into three subfamilies according to phylogenetic analyses. Evolutionary analysis indicated that multiple duplication events play important role in the expansion of the CUL gene family in Rosaceae. The expression pattern shows that only four CUL genes were highly expressed in pollen and pollen tube in pear. Of these CUL genes, PbCUL1.C1 presented the highest levels of expression in pollen, but the knockdown of PbCUL1.C1 by antisense oligonucleotide (ASO) did not influence pollen tube growth. Interestingly, the pollen tube treated by both the non-self S-RNase and ASO of PbCUL1.C1 was longer than that treated by unique non-self S-RNase, suggesting that the PbCUL1.C1 plays an important role for pollen tube growth underlying non-self S-RNase in pear. These results will be useful for exploring the biological roles of CUL genes in self-incompatibility in Rosaceae trees.







Similar content being viewed by others
Abbreviations
- CUL :
-
Cullin
- ASO:
-
antisense oligonucleotide
- CRLs:
-
cullin-RING ligase complexes
- SLF:
-
S-locus F-box
- SCF:
-
S-phase kinase-associated protein 1 (SKP1)-CUL1 (CUL1)-F-box-RING box 1 (RBX1)
- HMM:
-
Hidden Markov model
- RT-PCR:
-
real-time PCR
- RNA-Seq:
-
RNA sequencing
- MP:
-
mature pollen grains
- HP:
-
hydrated pollen grains
- PT:
-
growing pollen tubes
- SPT:
-
stopped-growth pollen tubes
- qRT-PCR:
-
quantitative real-time PCR
- WGD:
-
whole-genome duplication
References
Benanti JA, Cheung SK, Brady MC, Toczyski DP (2007) A proteomic screen reveals SCFGrr1 targets that regulate the glycolytic-gluconeogenic switch. Nat Cell Biol 9(10):1184–1191
Brown PH, Ho TH (1986) Barley aleurone layers secrete a nuclease in response to gibberellic acid: purification and partial characterization of the associated ribonuclease, deoxyribonuclease, and 3′-nucleotidase activities. Plant Physiol 82:801–806
Chen G, Zhang B, Zhao Z, Sui Z, Zhang H, Xue Y (2010) ‘A life or death decision’ for pollen tubes in S-RNase-based self-incompatibility. J Exp Bot 61:2027–2037
Choi CM, Gray WM, Mooney S, Hellmann H (2015) Composition, roles, and regulation of ccullin-based ubiquitin E3 ligases. Arabidopsis Book 12:e0175
Dai C, Xue HW (2010) Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J 29(11):1916–1927
Devoto A, Nieto-Rostro M, Xie DX, Ellis C, Harmston R, Patrick E, Davis J, Sherratt L, Coleman M, Turner JG (2002) COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J 32(4):457–466
De Nettancourt D (2001) Incompatibility and incongruity in wild and cultivated plants, 2nd edn. Springer, Heidelberg
Dong M, Zhaoyu G, Wei L, Aide W, Hui Y, Qing Y, Tianzhong L (2014) Apple MdABCF assists in the transportation of S-RNase into pollen tubes. Plant J 78:990–1002
Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14:755–763
Franklin-Tong VE (2008) Self-incompatibility in flowering plants. Springer, Heidelberg
Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci U S A 99:11519–11524
Gomi K, Sasaki A, Itoh H, Ueguchi-Tanaka M, Ashikari M, Kitano H, Matsuoka M (2004) GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J 37(4):626–634
Graaf BHJD, Cheung AY, Andreyeva T, Levasseur K, Kieliszewski M, Wu H (2005) Rab11 GTPase-regulated membrane trafficking is crucial for tip-focused pollen tube growth in Tobacco. Plant Cell 17:2564–2579
Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M (1999) Identification of an SCF ubiquitin–ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev 13(13):1678–1691
Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414(6861):271–276
Guo L, Nezames CD, Sheng L, Deng X, Wei N (2013) Cullin-RING ubiquitin ligase family in plant abiotic stress pathways. J Integr Plant Biol 55:21–30
Hershko A (1998) The ubiquitin system. Annu Rev Biochem 67:425–479
Hotton SK, Callis J (2008) Regulation of cullin RING ligases. Annu Rev Plant Biol 59:467–489
Hua Z, Kao TH (2006) Identification and characterization of components of a putative Petunia S-locus F-box-containing E3 ligase complex involved in S-RNase-based self-incompatibility. Plant Cell 18:2531–2553
Hua Z, Vierstra RD (2011) The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol 62:299–334
Initiative AG (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Iwano M, Takayama S (2012) Self/non-self discrimination in angiosperm self-incompatibility. Curr Opin Plant Biol 15:78–83
Jia FJ, Wang CY, Huang JG, Yang GD, Wu CG, Zheng CC (2015) SCF E3 ligase PP2-B11 plays a positive role in response to salt stress in Arabidopsis. J Exp Bot 66(15):4683–4697
Jimenez-Duran K, Mcclure BA, Garcia-Campusano F, Rodriguez-Sotres R, Cisneros J, Busot G, Cruz-Garcia F (2013) NaStEP: a proteinase inhibitor essential to self-incompatibility and a positive regulator of HT-B stability in Nicotiana alata pollen tubes. Plant Physiol 161:97–107
Ken-Ichi K et al (2010) Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science 330:796–799
Kipreos et al (1996) Cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell 85:829–839
Konopka-Postupolska D, Clark G, Goch G, Debski J, Floras K, Cantero A, Fijolek B, Roux S, Hennig J (2009) The role of Annexin 1 in drought stress in Arabidopsis. Plant Physiol 150(3):1394–1410
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645
Kubo K, Paape T, Hatakeyama M, Entani T, Takara A, Kajihara K, Tsukahara M, Shimizu-Inatsugi R, Shimizu KK, Takayama S (2015) Gene duplication and genetic exchange drive the evolution of S-RNase-based self-incompatibility in Petunia. Nature Plants 1:14005
Kubo KI, Tsukahara M, Fujii S, Murase K, Wada Y, Entani T, Iwano M, Takayama S (2016) Cullin1-P is an essential component of non-self recognition system in self-incompatibility in Petunia. Plant Cell Physiol 57(11):2403–2416
Lan Z, Jian H, Zhonghua Z, Qun L, Sims TL, Yongbiao X (2010) The Skp1-like protein SSK1 is required for cross-pollen compatibility in S-RNase-based self-incompatibility. Plant J 62:52–63
Li S, Sun P, Williams JS, Kao TH (2014) Identification of the self-incompatibility locus F-box protein-containing complex in Petunia inflate. Plant Reprod 27:31–45
Liao F, Wang L, Yang LB, Zhang L, Peng X, Sun MX (2013) Antisense oligodeoxynucleotide inhibition as an alternative and convenient method for gene function analysis in pollen tubes. PLoS One 8:e59112
Luu DT, Qin X, Morse D, Cappadocia M (2000) S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibility. Nature 407:649–651
Mcclure B, Mou B, Canevascini S, Bernatzky R (1999) A small asparagine-rich protein required for S-allele-specific pollen rejection in Nicotiana. P Natl Acad Sci USA 96:13548–13553
McClure BA, Gray JE, Anderson MA, Clarke AE (1990) Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature 347:757–760
Minamikawa MF, Koyano R, Kikuchi S, Koba T, Sassa H (2014) Identification of SFBB-containing canonical and noncanonical SCF complexes in pollen of apple (Malus × domestica). PLoS One 9:e97642
Sarikas A, Hartmann T, Pan ZQ (2011) The cullin protein family. Genome Biol 12:220
Sassa H, Hirano H, Ikehashi H (1992) Self-incompatibility-related RNases in styles of Japanese pear (Pyrus serotina Rehd.). Plant Cell Physiol 33:811–814
Verde I, Abbott AG, Scalabrin S et al (2013) The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet 45:487–494
Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40:e49–e49
Wang Y, Tsukamoto T, K-w Y, Wang X, Huang S, McCubbin AG, T-h K (2004) Chromosome walking in the Petunia inflata self-incompatibility (S-) locus and gene identification in an 881-kb contig containing S2-RNase. Plant Mol Biol 54:727–742
Wentao L, Chetelat RT (2010) A pollen factor linking inter- and intraspecific pollen rejection in tomato. Science 330:1827–1830
Wu J, Wang Z, Shi Z, Zhang S, Ming R, Zhu S, Khan MA, Tao S, Korban SS, Wang H, Chen NJ, Nishio T, Xu X, Cong L, Qi K, Huang X, Wang Y, Zhao X, Wu J, Deng C, Gou C, Zhou W, Yin H, Qin G, Sha Y, Tao Y, Chen H, Yang Y, Song Y, Zhan D, Wang J, Li L, Dai M, Gu C, Wang Y, Shi D, Wang X, Zhang H, Zeng L, Zheng D, Wang C, Chen M, Wang G, Xie L, Sovero V, Sha S, Huang W, Zhang S, Zhang M, Sun J, Xu L, Li Y, Liu X, Li Q, Shen J, Wang J, Paull RE, Bennetzen JL, Wang J, Zhang S (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 23:396–408
Xu C, Li M, Wu J, Guo H, Li Q, Zhang Y’, Chai J, Li T, Xue Y (2013) Identification of a canonical SCFSLF complex involved in S-RNase-based self-incompatibility of Pyrus (Rosaceae). Plant Mol Biol 81:245–257
Yuan H, Meng D, Gu Z, Li W, Wang A, Yang Q, Zhu Y, Li T (2014) A novel gene, MdSSK1, as a component of the SCF complex rather than MdSBP1 can mediate the ubiquitination of S-RNase in apple. J Exp Bot 65:3121–3131
Zhang S-L, Hiratsuka S (2000) Cultivar and developmental differences in S-protein concentration and self-incompatibility in the Japanese pear. HortScience 35:917–920
Zhao L, Ma W, Han B, Liang L, Zhang Y, Hong G, Xue Y (2002) An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol Bio 50:29–42
Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP (2002) Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature 416:703–719
Zhou H, Yin H, Chen J, Liu X, Gao Y, Wu J, Zhang S (2016) Gene-expression profile of developing pollen tube of Pyrus bretschneideri. Gene Expr Patterns 20:11–21
Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, Miller DJ, Walden H, Duda DM, Seyedin SN, Hoggard T, Harper JW, White KP, Schulman BA (2009) Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell 36:39–50
Funding
This work was financially supported by the National Natural Science Foundations of China (31772276 and 31672118).
Author information
Authors and Affiliations
Contributions
SLZ and CG conceived and designed the experiments. YZ wrote the manuscript. YZ, YDH, LW, GMW conducted all the experiments and data analysis. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key Message
1. Fifty-seven CUL genes were identified from pear and other rosaceae fruit trees and can be phylogenetic classified into three groups;
2. Pear PbCUL1.C1 is expressed in all the tested tissues and more highly expressed in pollen tube;
3. The down-regulation of PbCUL1.C1 enhances pollen tube growth underlying non-self S-RNase.
Electronic supplementary material
Fig S1
The yeast two-hybrid assay. Full-length sequences of PbCUL1.C1 were amplified and inserted into vector PGBKT7. Full-length sequences of PbSSK1 and PbSSK2 were amplified and inserted into vector PGADT7. The LiC1-PEG method was used to co-transform the vectors into yeast strain AH109 cells. The transformants were selected on SD/Leu/-Trp medium. The interaction was detected on SD/-Leu/-Trp/-His/-Ade medium. The transformation of PGADT7-T and PGBKT7-Lam, PGADT7-T, and PGBKT7-53 were used as negative and positive controls. (PDF 487 kb)
Table S1
Primer pairs for CUL genes used for quantitative real-time PCR analysis. (XLSX 11 kb)
Table S2
The primer sequences used for ASO. (XLSX 10 kb)
Rights and permissions
About this article
Cite this article
Zhou, Y., Huang, Y., Wu, L. et al. Phylogenetic and Expression Analyses of Cullin Family Members Unveil the Role of PbCUL1.C1 in Pollen Tube Growth Underlying Non-self S-RNase in Pear. Plant Mol Biol Rep 38, 601–612 (2020). https://doi.org/10.1007/s11105-020-01221-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-020-01221-2