Abstract
Osteosarcoma is a type of aggressive malignant bone tumour that frequently metastasizes to lungs, resulting in poor prognosis. However, the molecular mechanisms of lung metastasis of osteosarcoma remain poorly understood. Here we identify exon–intron fusion genes in osteosarcoma cell lines and tissues. These fusion genes are derived from chromosomal translocations that juxtapose the coding region for amino acids 1–38 of Rab22a (Rab22a1–38) with multiple inverted introns and untranslated regions of chromosome 20. The resulting translation products, designated Rab22a-NeoFs, acquire the ability to drive lung metastasis of osteosarcoma. The Rab22a1–38 moiety governs the function of Rab22a-NeoFs by binding to SmgGDS-607, a GTP–GDP exchange factor of RhoA. This association facilitates the release of GTP-bound RhoA from SmgGDS-607, which induces increased activity of RhoA and promotes metastasis. Disrupting the interaction between Rab22a-NeoF1 and SmgGDS-607 with a synthetic peptide prevents lung metastasis in an orthotopic model of osteosarcoma. Our findings may provide a promising strategy for a subset of osteosarcoma patients with lung metastases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
RNA-seq and WGS data that support the findings of this study have been deposited in the Sequence Read Archive (SRA) under the accession code SRP181860. The human RAB22a fusion data were obtained from the The Cancer Genome Atlas Research Network (http://cancergenome.nih.gov/) and cBio portal (http://www.cbioportal.org/). RNA22A point-mutation data were retrieved from Catalogue Of Somatic Mutations In Cancer (https://cancer.sanger.ac.uk/cosmic/) and the cBio portal database. Source data for Figs. 1–8 and Extended Data Figs. 1–10 are available online. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Change history
05 February 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41556-024-01370-6
11 June 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41556-020-0541-9
References
Rosenberg, A. E. WHO classification of soft tissue and bone, fourth edition: summary and commentary. Curr. Opin. Oncol. 25, 571–573 (2013).
Mirabello, L., Troisi, R. J. & Savage, S. A. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer 115, 1531–1543 (2009).
Dorfman, H. D. & Czerniak, B. Bone cancers. Cancer 75, 203–210 (1995).
Valery, P. C., Laversanne, M. & Bray, F. Bone cancer incidence by morphological subtype: a global assessment. Cancer Causes Control 26, 1127–1139 (2015).
Zambo, I. & Vesely, K. WHO classification of tumours of soft tissue and bone 2013: the main changes compared to the 3rd edition. Cesk. Patol. 50, 64–70 (2014).
Lorenz, S. et al. Unscrambling the genomic chaos of osteosarcoma reveals extensive transcript fusion, recurrent rearrangements and frequent novel TP53 aberrations. Oncotarget 7, 5273–5288 (2016).
Kovac, M. et al. Exome sequencing of osteosarcoma reveals mutation signatures reminiscent of BRCA deficiency. Nat. Commun. 6, 8940 (2015).
Stephens, P. J. et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40 (2011).
Chen, X. et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 7, 104–112 (2014).
Kansara, M., Teng, M. W., Smyth, M. J. & Thomas, D. M. Translational biology of osteosarcoma. Nat. Rev. Cancer 14, 722–735 (2014).
Perry, J. A. et al. Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc. Natl Acad. Sci. USA 111, E5564–E5573 (2014).
Helman, L. J. & Meltzer, P. Mechanisms of sarcoma development. Nat. Rev. Cancer 3, 685–694 (2003).
Gokgoz, N. et al. Comparison of p53 mutations in patients with localized osteosarcoma and metastatic osteosarcoma. Cancer 92, 2181–2189 (2001).
Ji, J. et al. Inherited germline ATRX mutation in two brothers with ATR-X syndrome and osteosarcoma. Am. J. Med. Genet. A 173, 1390–1395 (2017).
Smolle, M. A. et al. A novel mutation in ATRX associated with intellectual disability, syndromic features, and osteosarcoma. Pediatr. Blood Cancer 64, 26522 (2017).
Mertens, F., Johansson, B., Fioretos, T. & Mitelman, F. The emerging complexity of gene fusions in cancer. Nat. Rev. Cancer 15, 371–381 (2015).
Drilon, A. et al. Entrectinib in ROS1 fusion-positive non-small-cell lung cancer: integrated analysis of three phase 1–2 trials. Lancet Oncol. 21, 261–270 (2020).
Doebele, R. C. et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1–2 trials. Lancet Oncol. 21, 271–282 (2020).
Brien, G. L., Stegmaier, K. & Armstrong, S. A. Targeting chromatin complexes in fusion protein-driven malignancies. Nat. Rev. Cancer 19, 255–269 (2019).
Latysheva, N. S. & Babu, M. M. Discovering and understanding oncogenic gene fusions through data intensive computational approaches. Nucleic Acids Res. 44, 4487–4503 (2016).
Tomlins, S. A. et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature 448, 595–599 (2007).
Singh, D. et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 337, 1231–1235 (2012).
Zou, C. Y. et al. Establishment and characteristics of two syngeneic human osteosarcoma cell lines from primary tumor and skip metastases. Acta Pharmacol. Sin. 29, 325–332 (2008).
Tang, Q. L. et al. Glycogen synthase kinase-3β, NF-κB signaling, and tumorigenesis of human osteosarcoma. J. Natl Cancer Inst. 104, 749–763 (2012).
Yin, J. Q. et al. Bufalin induces apoptosis in human osteosarcoma U-2OS and U-2OS methotrexate300-resistant cell lines. Acta Pharmacol. Sin. 28, 712–720 (2007).
Anderson, N. D. et al. Rearrangement bursts generate canonical gene fusions in bone and soft tissue tumors. Science 361, eaam8419 (2018).
Shen, M. M. Chromoplexy: a new category of complex rearrangements in the cancer genome. Cancer Cell 23, 567–569 (2013).
Chen, G. et al. MRI-visible polymeric vector bearing CD3 single chain antibody for gene delivery to T cells for immunosuppression. Biomaterials 30, 1962–1970 (2009).
Mao, S. et al. Synthesis, characterization and cytotoxicity of poly(ethylene glycol)-graft-trimethyl chitosan block copolymers. Biomaterials 26, 6343–6356 (2005).
Kamai, T. et al. Overexpression of RhoA, Rac1, and Cdc42 GTPases is associated with progression in testicular cancer. Clin. Cancer Res. 10, 4799–4805 (2004).
Sahai, E. & Marshall, C. J. RHO-GTPases and cancer. Nat. Rev. Cancer 2, 133–142 (2002).
Hamel, B. et al. SmgGDS is a guanine nucleotide exchange factor that specifically activates RhoA and RhoC. J. Biol. Chem. 286, 12141–12148 (2011).
Shimizu, H. et al. Structure-based analysis of the guanine nucleotide exchange factor SmgGDS reveals armadillo-repeat motifs and key regions for activity and GTPase binding. J. Biol. Chem. 292, 13441–13448 (2017).
Jennings, B. C., Lawton, A. J., Rizk, Z. & Fierke, C. A. SmgGDS-607 regulation of RhoA GTPase prenylation is nucleotide-dependent. Biochemistry 57, 4289–4298 (2018).
Eathiraj, S., Pan, X., Ritacco, C. & Lambright, D. G. Structural basis of family-wide Rab GTPase recognition by rabenosyn-5. Nature 436, 415–419 (2005).
Sugahara, K. N. et al. Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer Cell 16, 510–520 (2009).
Sugahara, K. N. et al. Coadministration of a tumor-penetrating peptide enhances the efficacy of cancer drugs. Science 328, 1031–1035 (2010).
Garcia-Torres, D. & Fierke, C. A. The chaperone SmgGDS-607 has a dual role, both activating and inhibiting farnesylation of small GTPases. J. Biol. Chem. 294, 11793–11804 (2019).
Shimizu, H., Toma-Fukai, S., Kontani, K., Katada, T. & Shimizu, T. GEF mechanism revealed by the structure of SmgGDS-558 and farnesylated RhoA complex and its implication for a chaperone mechanism. Proc. Natl Acad. Sci. USA 115, 9563–9568 (2018).
Berg, T. J. et al. Splice variants of SmgGDS control small GTPase prenylation and membrane localization. J. Biol. Chem. 285, 35255–35266 (2010).
Latysheva, N. S. et al. Molecular principles of gene fusion mediated rewiring of protein interaction networks in cancer. Mol. Cell 63, 579–592 (2016).
Kim, D. & Salzberg, S. L. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 12, R72 (2011).
Ma, C., Shao, M. & Kingsford, C. SQUID: transcriptomic structural variation detection from RNA-seq. Genome Biol. 19, 52 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Tarasov, A., Vilella, A. J., Cuppen, E., Nijman, I. J. & Prins, P. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015).
Liao, D. et al. Aspirin suppresses the growth and metastasis of osteosarcoma through the NF-κB pathway. Clin. Cancer Res. 21, 5349–5359 (2015).
Maddika, S. et al. WWP2 is an E3 ubiquitin ligase for PTEN. Nat. Cell Biol. 13, 728–733 (2011).
Ren, X. D. & Schwartz, M. A. Determination of GTP loading on Rho. Methods Enzymol. 325, 264–272 (2000).
Wang, X. et al. Targeting the CK1α/CBX4 axis for metastasis in osteosarcoma. Nat. Commun. 11, 1141 (2020).
American Association for Cancer Research. Protective effect of aspirin associated with SNP. Cancer Discov. https://doi.org/10.1158/2159-8290.CD-NB2014-011 (2014).
Acknowledgements
We thank X. Chen (St Jude Children’s Research Hospital) for his help with analysis of WGS and RNA-seq data. This work was supported by the National Key Research and Development Program of China 2016YFA0500304 to T.K. and 2016YFA0503100 to Jian Chen, the National Nature Science Foundation in China (NSFC) 81502512 to D.L., 81902738 to L.Z. and 81530081 to T.K. and Science and Technology Program of Guangzhou, China (grant no. 201508020102 and 201607020038 to T.K.).
Author information
Authors and Affiliations
Contributions
T.K. conceived the idea. D.L., L. Z. and C. Z. performed most experiments. L.Z. and Jinna Chen performed the FISH. X.H. and J.S. generated the monoclonal antibodies (RAD5-8 and hRAD5-8-v1-R5). X.S. generated the nanoparticles. J.Y. and J.-N.S. collected clinical samples. H.Z. and S.G. analysed the structures and purified proteins. D.L., L.Z., J.Y., X.W., R.Z., X.-Y.G., W.D., Y.-X.Z., Jian Chen and T.K. analysed the data. D.L., S.G., Jian Chen and T.K. wrote the manuscript. All co-authors have seen and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Identification of the chromosomal translocation-derived aberrant RAB22A as the driver for osteosarcoma lung metastasis.
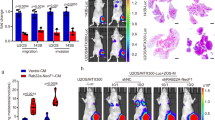
a, RT-PCR (left panels) for each RAB22A-NeoF1-6 was performed as indicated, followed by Sanger sequencing using the indicated cell lines. The reading frames at the breakpoints are shown by the Sanger sequencing chromatogram (right panels). Data are representatives of n =3 biologically independent experiments. b, c, The whole-genome sequencing of ZOS and ZOS-M revealed that a region of chromosome 20 was also associated with chromosome chromothripsis. d, g, Quantification analyses of cell viability by the MTT assay in the indicated stable cells at 24h. Mean ± s.d. of n = 3 biologically independent experiments. p values are shown. Two-tailed Student t-test. Vector, vector-only control. e, f, Quantification analyses of the migration (e) and invasion (f) assays using the indicated stable cell lines. Mean ± s.d. of n = 4 biologically independent experiments. p values are shown. Two-tailed Student t-test. Vector, vector-only control. h, Quantification analyses of wet lung weight from the nude mice in Fig. 1e. Mean ± s.d. of n = 6 biologically independent animals. p values are shown. Two-tailed Student t-test. Vector, vector-only control. i, j, Kaplan-Meier survival data were plotted as the percent of mice surviving in the indicated group using a predefined cut off tumor volume of 1.5 cm3. n=6 biologically independent animals. p values are shown. The Log-rank test was performed. Vector, vector-only control. Statistical source data are provided in Source Data Extended Data Fig. 1.
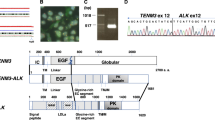
Extended Data Fig. 2 RAB22A-NeoF1 is dominant in ZOS and ZOS-M cells.
a, The protein levels of each RAB22A-NeoF1-6 in U2OS cells transiently transfected with the indicated plasmids with or without 200 nm Bafilomycin A1 and 10 μm MG132 for 6 h. Vector, vector-only control. b, The reads of RAB22A-NeoF1-6 in both ZOS and ZOS-M cell lines. c, The detection of endogenous RAB22A-NeoF2/3/4/5/6 proteins by Western blotting in the indicated stable cell lines, as described in methods (Note: The Western blotting using anti-RAB22A-N followed by IP with anti-RAB22A-N was performed with the cut membranes below the molecular weight of 15 KDa to avoid the effects of wild type RAB22A). Data in a,c are representatives of n =3 biologically independent experiments. sgNC, negative control sgRNA. d, The amino acid sequence of RAB22A-NeoF1 full-length protein. Residues corresponding to the inverse DOK5 sequences are shown in red. e, Quantification analysis of mRNA levels of RAB22A-NeoF1 in the indicated osteosarcoma cell lines by real-time PCR. Mean ± s.d. of n = 3 biologically independent experiments. p values are shown. Two-tailed Student t-test. f, FISH analyses in ZOS and ZOS-M cells, as described in the methods. Red: RAB22A, Green: DOK5, yellow: the fusion of RAB22A and DOK5 indicated by arrows. Data are representatives of n = 3 biologically independent experiments. Statistical source data are provided in Source Data Extended Data Fig. 2.
Extended Data Fig. 3 RAB22A-NeoF1 promotes osteosarcoma lung metastasis.
a, Binding specificity of mAb RAD5-8 against RAB22A-NeoF1 fusion protein. 293T cells transfected with the indicated plasmids or vector were lysed and mixed with 2xSDS sample buffer (Left panel), and the purified RAB22A-NeoF1-hFc fusion protein (Right panel), were then analyzed by Western blotting by mAb RAD5-8. Data are representatives of n = 3 biologically independent experiments. b, The effects of shRNA against RAB22A-NeoF1 detected by qRT-PCR. shNC, negative control shRNA. c, d, Quantification analyses of migration and invasion assays using the indicated stable cell lines. NC, negative control shRNA. e, Detection of RAB22A-NeoF1 in the indicated stable cell lines by Western blotting. Data are representatives of n = 3 biologically independent experiments. Vector, vector-only control. f, Quantification analyses of migration and invasion assays using the indicated stable cell lines. Data in b-d, f are mean ± s.d. of n=3 biologically independent experiments, p values are shown. Two-tailed Student’s t-test. Vector, vector-only control. g–k, The orthotopic osteosarcoma metastasis model in vivo using the indicated stable cell lines (n=6 biologically independent animals). g, i, Quantification analyses of wet lung weights. Data are the mean ± s.d. p values are shown. A two-tailed Student’s t-test was performed. h–k, Kaplan-Meier survival data were plotted as the percent of mice surviving in the indicated groups using a predefined cutoff tumor volume of 1.5 cm3. p values are shown. The Log-rank test was performed. NC, negative control shRNA. Vector, vector-only control. l, m, Representative images of mice from the tail vein injection metastasis model in vivo using the indicated stable cell lines (n=6 biologically independent animals). NC, negative control shRNA. Vector, vector-only control. Statistical source data are provided in Source Data Extended Data Fig. 3.
Extended Data Fig. 4 RAB22A-NeoF1 specifically activates RhoA, and depletion of RhoA abolishes the functions of RAB22A-NeoF1 in vitro.
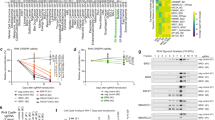
a, The partial list of proteins from the mass spectrum using cells stably expressing RAB22A-NeoF1-SFB. The unique and total peptide numbers for the indicated proteins are shown. b, c, e, 293T cells co-transfected with the indicated plasmids for 48 h were subjected to the indicated assays. d, The detection of Rho-GTP and membrane RhoA (RhoA-MEM) in the indicated stable cell lines by Western blotting. Vector, vector-only control. f, The co-IPs were performed using ZOS cells with IgG, mAb RAD5-8 or anti-RhoA at their endogenous levels, and were analyzed by Western blotting using hRAD5-8-V1-R5. g, The indicated proteins were analyzed by Western blotting in the indicated stable cells. Data in b-g are representatives of n = 3 biologically independent experiments. h, i, Quantification analyses of the migration and invasion (h) or adhesion (i) assays using the indicated stable cell lines. Vector, vector-only control. j, k, Quantification analyses of the migration and invasion (j) or adhesion (k) assays using the indicated stable cell lines with or without the RhoA inhibitor CT04. Data in h-k are Mean ± s.d. of n = 3 biologically independent experiments. p values are shown. A two-tailed Student’s t-test was performed. Vector, vector-only control. Statistical source data are provided in Source Data Extended Data Fig. 4.
Extended Data Fig. 5 RAB22A-NeoF1 promotes osteosarcoma lung metastasis in a manner dependent on RhoA.
a, SgRNA1, 2-mediated knockout of RhoA in RAB22A-NeoF1 overexpression cells with or without re-introduction of RhoA back were analyzed by Western blotting. Data is representative of n = 3 biologically independent experiments. NC, negative control sgRNA. b, c, Quantification analyses of the migration (b) and invasion (c) assays using the indicated stable cell lines. Mean ± s.d. of n = 3 biologically independent experiments. p values are shown. Two-tailed Student t-test. NC, negative control sgRNA. Vector, vector-only control. d–f, The orthotopic osteosarcoma metastasis model in vivo using the indicated stable cell lines (n=6 biologically independent animals). d, Quantification analyses of wet lung weights. Data are the mean ± s.d. p values are shown. Two-tailed Student’s t-test. e, f, Kaplan-Meier survival data were plotted as the percent of mice surviving in the indicated group using a predefined cutoff tumor volume of 1.5 cm3. p values are shown. The Log-rank test was performed. NC, negative control shRNA. Vector, vector-only control. g, Representative images of mice from the tail vein injection metastasis model in vivo using the indicated stable cell lines (n=6 biologically independent animals). NC, negative control shRNA. Vector, vector-only control. Statistical source data are provided in Source Data Extended Data Fig. 5.
Extended Data Fig. 6 RAB22A-NeoF1 promotes osteosarcoma lung metastasis through interacting with SmgGDS607.
a, Detection of SmgGDS in osteosarcoma cell lines by Western blotting. b, The effects of shRNA specially against the SmgGDS558 (BD), SmgGDS607 (C1) or both (I1, I2) in the indicated cells, as detected by Western blotting. NC, negative control shRNA. c, 293T cells co-transfected with the indicated plasmids for 48 h were subjected to the indicated assays. d, The co-IPs were performed using ZOS cells with IgG, mAb RAD5-8 or anti-SmgGDS at their endogenous levels, and were analyzed by Western blotting using hRAD5-8-V1-R5. e, SgRNA1, 2-mediated knockout of SmgGDS in RAB22A-NeoF1 overexpression cells with or without re-introduction of wild-type SmgGDS607 or its E254K+D256K mutant back were analyzed by Western blotting. Data in a-e are representatives of n = 3 biologically independent experiments. sgNC, negative control sgRNA. Vector, vector-only control. f, Quantification analyses of adhesion of the indicated cells used in e. mean ± s.d. of n=3 independent biological experiments. p values are shown. Two-tailed Student’s t-test. g–j, The orthotopic osteosarcoma metastasis model in vivo using the indicated stable cell lines used in e (n=6 biologically independent animals). g, i, Kaplan-Meier survival data were plotted as the percent of mice surviving in the indicated groups using a predefined cutoff tumor volume of 1.5 cm3. p values are shown. The Log-rank test was performed. h, j, Quantification analyses of wet lung weights. Data are the mean ± s.d. p values are shown. Two-tailed Student’s t-test. sgNC, negative control sgRNA. Vector, vector-only control. Statistical source data are provided in Source Data Extended Data Fig. 6.
Extended Data Fig. 7 SmgGDS607 is also crucial in lung metastasis of ZOS-M cells that have endogenous RAB22A-NeoF1.
a, The detection of endogenous SmgGDS protein by Western blotting in the indicated ZOS-M stable cell lines. The experiments were repeated three times independently with similar results. NC, negative control sgRNA. b, c, Quantification analyses of adhesion (b) migration and invasion (c) assays using the indicated stable cell lines. mean ± s.d. of n=3 independent biological experiments. p values are shown. Two-tailed Student’s t-test. sgNC, negative control sgRNA. d–g, The orthotopic osteosarcoma metastasis model in vivo using the indicated stable cell lines (n=6 biologically independent animals). d, Representative images of mice. e, H&E staining of the lungs from representative tumor-bearing nude mice. Scale bars, 5mm. f, Quantification analyses of lung nodules. g, Quantification analyses of wet lung weights. f, g, Data are the mean ± s.d. p values are shown. A two-tailed Student’s t-test was performed. sgNC, negative control sgRNA. Statistical source data are provided in Source Data Extended Data Fig. 7.
Extended Data Fig. 8 Amino acids 1-10 of RAB22A-NeoF1 are responsible for its binding to SmgGDS607.
a, The fragments of RAB22A-NeoF1. b, d, 293T cells co-transfected with the indicated plasmids for 48 h were subjected to the indicated assays. c, 293T cells transfected with the indicated plasmids were lysed and subjected to immunoprecipitation using streptomycin beads and then eluted with biotin. The eluates were incubated with the purified SmgGDS607-His protein and the immunoprecipitated complex using a nickel column followed by Western blotting. Vector, vector-only control. e, f, Detection of RhoA-GTP and membrane RhoA (RhoA-MEM) in the indicated stable cell lines by Western blotting. Data in b-f are representatives of n = 3 biologically independent experiments. Vector, vector-only control.
Extended Data Fig. 9 Both Arg4 and Lys7 of RAB22A-NeoF1 are required for the functions of RAB22A-NeoF1.
a, g, Quantification analyses of adhesion of the indicated cells. Mean ± s.d. of n = 3 biologically independent experiments, p values are shown. Two-tailed Student’s t-test. Vector, vector-only control. b–d, h–j, The orthotopic osteosarcoma metastasis model in vivo using the indicated stable cell lines (n=6 biologically independent animals). b, h, Quantification analyses of wet lung weights. Data are the mean ± s.d. p values are shown. A two-tailed Student’s t-test was performed. c, d, i, j, Kaplan-Meier survival data were plotted as the percent of mice surviving in the indicated groups using a predefined cutoff tumor volume of 1.5 cm3. p values are shown. The Log-rank test was performed. Vector, vector-only control. e, Ambience of key residues of R4 and K7 in RAB22A. RAB22A is shown as a cartoon representation with residues 1-10 colored cyan. R4, K7, and the residues surrounding K7 are shown as ball-and-stick models. f, 293T cells transfected with the indicated plasmids were lysed and subjected to immunoprecipitation using streptomycin beads and then eluted with biotin. The eluates were incubated with the purified SmgGDS607-His protein and the immunoprecipitated complex using a nickel column followed by Western blotting. Data are representatives of n = 3 biologically independent experiments. Statistical source data are provided in Source Data Extended Data Fig. 9.
Extended Data Fig. 10 Blocking the interaction between RAB22A-NeoF1 and SmgGDS607 inhibits osteosarcoma lung metastasis.
a, Biotin-labeled peptide-WT or -DD was incubated with the different indicated doses of ZOS and ZOS-M cell lysates and pulled-down, and the eluates were subjected to Western blotting. b, The expression of ITGAV and NRP1 in the indicated osteosarcoma cell lines by Western blotting. Data in a, b were representatives of n = 3 biologically independent experiments. c–f, Quantification analyses of the migration (c, e) and invasion (d, f) assays using the indicated stable cell lines with or without WT–iRGD or DD-iRGD treatment at the indicated concentrations. Mean ± s.d. of n = 3 biologically independent experiments. p values are shown. Two-tailed Student t-test. Vector, vector-only control. g, Quantification analyses of the migration and invasion assays using the ZOS and ZOS-M cell lines with or without 10 μM of WT–iRGD or DD-iRGD treatment. Mean ± s.d. of n = 4 biologically independent experiments. p values are shown. Two-tailed Student t-test. control, without peptide treatment as the control. h–k, The orthotopic osteosarcoma metastasis model in vivo using 143B-Luc cells stably expressing RAB22A-NeoF1 (i, j. n=8 biologically independent animals) or ZOS-M-Luc cells (k. n=6 biologically independent animals) with or without WT-iRGD or DD-iRGD treatment at 1 mg/ml and 10 mg/ml. h, The procedure for this experiment. i, k, Quantification analyses of wet lung weight. i, k, Mean ± s.d. p values are shown. Two-tailed Student’s t-test. j, Kaplan-Meier survival data were plotted as the percent of mice surviving in the indicated groups using a predefined cutoff tumor volume of 1.5 cm3. p values are shown. The Log-rank test was performed. Vector, vector-only control. Statistical source data are provided in Source Data Extended Data Fig. 10.
Supplementary information
Supplementary Tables
Supplementary Tables 1–8
Source data
Source Data Fig. 1
Statistical source data
Source Data Fig. 1
Unprocessed gels
Source Data Fig. 2
Statistical source data
Source Data Fig. 2
Unprocessed western blots
Source Data Fig. 3
Statistical source data
Source Data Fig. 3
Unprocessed western blots
Source Data Fig. 4
Statistical source data
Source Data Fig. 4
Unprocessed western blots
Source Data Fig. 5
Statistical source data
Source Data Fig. 5
Unprocessed western blots
Source Data Fig. 6
Statistical source data
Source Data Fig. 6
Unprocessed western blots
Source Data Fig. 7
Statistical source data
Source Data Fig. 7
Unprocessed western blots
Source Data Fig. 8
Statistical source data
Source Data Extended Data Fig. 1
Statistical source data
Source Data Extended Data Fig. 1
Unprocessed gels
Source Data Extended Data Fig. 2
Statistical source data
Source Data Extended Data Fig. 2
Unprocessed western blots
Source Data Extended Data Fig. 3
Statistical source data
Source Data Extended Data Fig. 3
Unprocessed western blots
Source Data Extended Data Fig. 4
Statistical source data
Source Data Extended Data Fig. 4
Unprocessed western blots
Source Data Extended Data Fig. 5
Statistical source data
Source Data Extended Data Fig. 5
Unprocessed western blots
Source Data Extended Data Fig. 6
Statistical source data
Source Data Extended Data Fig. 6
Unprocessed western blots
Source Data Extended Data Fig. 7
Statistical source data
Source Data Extended Data Fig. 7
Unprocessed western blots
Source Data Extended Data Fig. 8
Unprocessed western blots
Source Data Extended Data Fig. 9
Statistical source data
Source Data Extended Data Fig. 9
Unprocessed western blots
Source Data Extended Data Fig. 10
Statistical source data
Source Data Extended Data Fig. 10
Unprocessed western blots
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liao, D., Zhong, L., Yin, J. et al. Chromosomal translocation-derived aberrant Rab22a drives metastasis of osteosarcoma. Nat Cell Biol 22, 868–881 (2020). https://doi.org/10.1038/s41556-020-0522-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-020-0522-z
This article is cited by
-
Protein lipidation in cancer: mechanisms, dysregulation and emerging drug targets
Nature Reviews Cancer (2024)
-
Insights into the complex interactions between Rab22a and extracellular vesicles in cancers
Inflammation Research (2024)
-
LRIG2 regulates cell proliferation, migration and apoptosis of osteosarcoma
BMC Cancer (2022)
-
Rab22a-NeoF1 fusion protein promotes osteosarcoma lung metastasis through its secretion into exosomes
Signal Transduction and Targeted Therapy (2021)
-
Rab22a-NeoF1: a promising target for osteosarcoma patients with lung metastasis
Signal Transduction and Targeted Therapy (2020)