Abstract
Autophagy, or cellular self-digestion, is an essential cellular process imperative for energy homeostasis, development, differentiation, and survival. However, the intrinsic factors that bring about the sex-biased differences in liver autophagy are still unknown. In this work, we found that autophagic genes variably expresses in the steroidogenic tissues, mostly abundant in liver, and is influenced by the individual’s sexuality. Starvation-induced autophagy in a time-dependent female-dominated manner, and upon starvation, a strong gender responsive circulating steroid-HK2 relation was observed, which highlighted the importance of estrogen in autophagy regulation. This was further confirmed by the enhanced or suppressed autophagy upon estrogen addition (male) or blockage (female), respectively. In addition, we found that estrogen proved to be the common denominator between stress management, glucose metabolism, and autophagic action in female fish. To understand further, we used estrogen receptor (ER)α- and ER-β2-knockout (KO) medaka and found ER-specific differences in sex-biased autophagy. Interestingly, starvation resulted in significantly elevated mTOR transcription (compared with control) in male ERα-KO fish while HK2 and ULK activation was greatly decreased in both KO fish in a female oriented fashion. Later, ChIP analysis confirmed that, NRF2, an upstream regulator of mTOR, only binds to ERα, while both ERα and ERβ2 are effectively pulled down the HK2 and LC3. FIHC data show that, in both ER-KO fish, LC3 nuclear-cytoplasmic transport and its associated pathways involving SIRT1 and DOR were greatly affected. Cumulatively, our data suggest that, ERα-KO strongly affected the early autophagic initiation and altered the LC3 nuclear-cytoplasmic translocation, thereby influencing the sex-biased final autophagosome formation in medaka. Thus, existence of steroid responsive autophagy regulatory-switches and sex-biased steroid/steroid receptor availability influences the gender-skewed autophagy. Expectedly, this study may furnish newer appreciation for gender-specific medicine research and therapeutics.
Similar content being viewed by others
Introduction
Autophagy, a sophisticatedly regulated catabolic process that mediates the lysosomal turnover of organelles and macromolecules, is strongly activated in stress conditions to ensure cell survival. It is stimulated above the basal or resting rate when nutrients are scarce, cells are under stress, and damaged organelles need to be degraded [1]. Regulated changes through autophagy provides a positive energetic balance and ensures renewal and proper functioning of the cell organelles [2,3,4]. Both constitutive and starvation mediated autophagy has been considered a promising target for disease therapies [5, 6] and has recently emerged as a mainstream research in the field of physiology and medicine [7]. The development of gender medicine, i.e., medicine taking into account the gender/sex differences in diagnosis and therapy of diseases, has opened newer arenas to investigate the physiological and pathophysiological distinction between the two genders, taking into factor the myriads of sex steroids involved, and thus, is a foot forward toward re-writing many chapters in modern medicine [8, 9].
Estrogen is a sex hormone which in addition to its essential role in sexual development and the reproductive system is also involved in various biological responses. In many cases, it is evident that estrogen has multiple effects on autophagy and plays an important role in promoting autophagy in several organs, including the liver [10, 11]. Estrogen signals choreographs the hepatic cell regeneration and differentiation via the two different types of receptors, namely, estrogen receptor-α (ERα) and -β (ERβ) [12, 13]. Studies have shown numerous effects of estrogenic action on autophagy and its importance in treating liver diseases like HCC, liver cirrhosis, etc. [14]. Owing to the tighter control of autophagy by estrogen, it would be imperative to determine the fluctuation of autophagy corresponding to the estrogenic and ER regulation under various physiological and pathological conditions in the liver [11]. Autophagy is crucial for energy homeostasis, development, differentiation, and survival, and although several autophagy related studies have been carried out in the liver, most of them are focused on the in vitro studies owing to the dynamic nature of the autophagic process. This opens up newer avenues to study the intrinsic factors that bring about the sex-biased differences in liver autophagy, in vivo.
Fish are widely recognized model due to their easy generation, handling, gender disparity, astonishing similarity human physiology, and comparatively less ethical issues [15]. Moreover, existence of autophagy has been reported in various fish [16,17,18]. So in the present work, using two evolutionarily distant fish, Japanese anchovy (Engraulis japonicas, budding marine fish model [19, 20]) and medaka (Oryzias latipes, established mammalian research model [15]), we confirmed the conserved autophagic regulation among fish and higher vertebrates, and then focused on to unravel the sex-biased liver autophagy. Further with the help of two ER knockouts (KO), we show that it is the estrogen-ERs-autophagy axis that bring about the sex-biased differences in the liver physiology.
Results
Liver autophagy is sex biased in fish
Liver is an important exo-endocrine organ in the body and displays various histo-morphological and transcriptional sexual differences [21]. Since our study depicted sexually dimorphic autophagic gene expression in the liver (Fig. 1a and S1A–D), we focused our attention on liver for further analysis. Autophagy responses and its associated genomics is generally conserved from yeast to mammals [22], and we also observed high evolutionary closeness of autophagy related genes (HK2, mTOR, AMPK, ULK1, Beclin1, LC3, etc.) from both Japanese anchovy and medaka with their mammalian homologs (Fig. S1E).
a Real-time PCR profiling of candidate autophagic genes. Brain (B); Gonad (G); Head kidney (HK); Liver (L) (N = 7). b LC3 puncta of medaka female and male hepatocytes, exposed to DMSO (vehicle control), Bafilomycin A1 (Baf-A1), and starvation, were photomicrographed (upper panel), analyzed using ImageJ software and plotted against respective groups (lower panel), N = 15 cells per group. White dotted lines were used to mark the cell boundaries. c, d TEM sections of adult female (N = 3) and male (N = 4) medaka showcasing the higher autophagy and lower vacuole formation in female than male. AL autolysosome; ER endoplasmic reticulum; L lysosome; M mitochondria; V vacuole. e Multicolor Fluorescent in situ hybridization (FISH) analysis of Japanese anchovy adult liver. Cellular localization of LC3 (green) and AMPK (red) were depicted in 6-day starved anchovy female, and the white arrow indicates the representative colocalized signal. The white box indicates the position of inset, and the inset (bounded in white box) shows the co-localization characteristics of LC3 and AMPK. Scale = 5 μm. f Ratio of circulating E (estrogen) and T (testosterone) plotted against respective liver hexokinase (HK2) transcription at different time points. Male (N = 4) and female (N = 4) data are plotted on Y1 and Y2 axis, respectively, with blue dotted line and red line. Note: Graphical data are represented as means ± SEM, and significance (p < 0.05) are denoted by different alphabets. See also Figs. S1 and S2.
Calcium ion homeostasis, apoptosis, glucose, and energy homeostasis are widely accepted modulators of autophagy. So, to comprehend the general autophagy mechanism in liver, we treated the Japanese anchovy liver fragments with various autophagy-altering chemicals under serum and glucose starvation, ex vivo. Liver fragment culture confirmed the interconnected network of calcium ion homeostasis, glucose and energy balance, apoptosis, and autophagy pathways (Fig. S1F). Further to unravel the autophagic sex bigotry in liver, we treated the Japanese anchovy male and female liver fragments with CaCl2 or nimodipine (calcium inhibitor) in combination with HK2 OV, under both serum and glucose starved condition. Excess addition of both calcium and hexokinase propelled autophagy (especially AMPK and LC3 fold activation) in a feministic way, thus highlighting the sexual variance in autophagic action (Figs. S1G and S1H). Later to confirm the sexual dimorphism in autophagy, we measured the LC3 puncta and performed Transmission Electron Microscopy (TEM) using medaka hepatocytes and liver fragments, respectively. We found that LC3 puncta was significantly higher in females (Fig. 1b). Similarly, TEM analysis also depicted several autophagosomes and autolysosome in females, while no such structures were observed in males (Fig. 1c, d). Contrastingly, the vacuoles were at a higher preponderance in males than their female counterparts (Fig. 1c, d).
Sexually dimorphic autophagic induction could be linked to several sex-sided processes, (i) energy homeostasis [23, 24] (ii) oxidative stress [25] and (iii) sex steroids [26]. Moreover, we recently found similar interconnection of stress, gluconeogenesis, and autophagy in the gonad [19]. To further explore the possibility of such interconnections, we conducted a starvation experiment using Japanese anchovy, and found that in vivo dietary restriction accelerated the autophagic gene expression and mitochondrial population, especially in the females (Fig. 1e, S1I–K, and S2A–E). Unlike the general notion where mitochondrial population supposedly decreases after starvation [27], this increased mitochondrial number might be attributed to energy limitation/scarcity which the cells tries to mitigate via mitochondrial fragmentation and increasing the ATP producing surfaces [28]. Notably, we also observed female tipped mitochondrial content. In this sense, 17β-estradiol and progesterone, highly produced in females and known inducer of autophagy [19], have been proved to be involved in enhancement of both mitochondrial respiratory chain related gene transcription and mitochondrial function [29,30,31] and thereby inducing female biased autophagy. Biochemically, starvation altered the glucose metabolism, glycogen storage, and oxidative stress parameters in a female biased manner (Fig. S2G). Since oxidative stress and nutrient status are some of the foremost regulators of autophagy, it is highly likely that the anti-chauvinism in autophagic action is a cumulative effect of female-dominated glucose regulation and stress response [25, 32].
Sex steroid modulates liver autophagy in fish
The amount of circulating estrogen (E) and testosterone (T) concentration in the body determines the steroidal difference between sexes, and additionally the E/T ratio (E:T) affects the body physiology [33]. Strong correlation was observed between the circulating E:T and HK2 concentration among different sexes (Fig. 1f and S2F), thereby illuminating the steroid-autophagy interconnection. Since the females showed a higher preponderance of both E content and autophagic response, henceforth, we concentrated on understanding the mechanism of estrogen regulated autophagy modulation in fish. Expectedly, E2 (17β-estradiol) addition or restriction not only influenced the energy equilibrium and ROS production, but also, respectively, induced or suppressed the male and female autophagic action as compared with their sexual counterparts (Fig. 2a, b and S2H). The mature females also exhibited higher autophagic gene transcription and estrogen concentration, and simultaneous lower testosterone amount than the immature females or mature and immature males (Fig. 2c–e and S2I–J) was observed. This furnishes the fact that the female supremacy in estrogen production/circulation has some intricate association with liver autophagic activity. Contextually, both E2 and progesterone are known to enhance the expression of ATG3, ATG5, BECN1 genes, and modulate the phosphorylation of AMPK, mTOR and Akt in mammary epithelial cells [26]. So, it is probably not the stress, but the actual circulating steroids that creates the discrepancies between the male and female reaction to stress, and this sex steroid regulated stress-effect accentuates the autophagic response in the body [34].
a Relative mRNA (to respective control) profiles of candidate autophagy related genes after AI (aromatase inhibitor, Upper) and E2 (lower) treatment (10 mg/kg feed) (N = 8). b Liver SOD (Superoxide dismutase) and serum glucose measurement. E2 treated XY and AI treated XX fish (N = 8) are plotted on Y1 (XY, blue) and Y2 (XX, red) axis. c Estrogen concentration and d HK2 mRNA transcription quantification in mature (M) and immature (Im) samples of each sex. e Pro-autophagic (HK2, LC3-I, and LC3-II) and anti-autophagic (mTOR) protein profiling done with western blotting, using Immature (N = 10), Mature (N = 9) and Spent (N = 4) male and female liver samples. GADPH was used as internal control. See also Fig. S2.
ER are predominant regulators of liver autophagy
Estrogen receptors (ERα, ERβ1 and ERβ2) are the cornerstone of estrogenic action in medaka [35]. Among the three isoforms, only ERα and ERβ2 display female-biased transcription in adult medaka liver [35]. To comprehend the intricacies of estrogen and autophagic inter-regulation, we generated null medaka tilling mutants of ERα and ERβ2 (Fig. 3a and S3A-C). Estrogen production and HK2 transcriptions were significantly reduced in the ER-KO (ER−/−) females, while no significant differences were observed in the ER−/− males (Fig. 3b, c). ChIP analysis revealed that medaka ERα, and not ERβ2, directly interacted with NRF2 (an upstream inhibitor of mTOR), while both ERs affected the HK2 and LC3 activities (Fig. 3d and S3D). ERα influenced the mTOR action, and expectedly, upon ERα KO, the NRF/mTOR activity either increased (females) or remained unaffected (males) (Fig. 3b). Starvation reduced the mTOR activity in all groups as compared with their respective fed counterparts, and the starved-ERα−/− males depicted significantly higher expression than the other groups (Fig. S3E). Thus, ERα might be responsible for antagonistically regulating the mTOR transcription in medaka liver. On the other hand, significant HK2 transcriptional suppression in ERβ2−/− female than the ERα−/− might be linked to the circulating steroid concentration (Fig. S3E). In a separate investigation, we observed that autophagic alterations in germ cells of ERβ2−/− females could be rescued by calcium signaling modulation [18]. In eukaryotes, AMPK-independent calcium influx-regulated autophagy has also been characterized [36]. Therefore, it is possible that ERα is instrumental in bringing forth the constitutive HK2/mTOR/AMPK mediated autophagy, while ERβ2 is essential for the non-canonical, probably mTOR/AMPK independent-Ca+2/regulated autophagy. Hence, the higher mTOR augmentation in ERα−/− males than the females, probably due to the male dominated mTOR phosphorylation, might have resulted in the subsequent alterations in P53 positive cells [37] (Fig. S3F). These facts suggest that maintenance of the threshold level of both estrogen and ERs is important for the initiation of the early autophagy factors.
a Phenotypic differences between wild, ERα−/−, and ERβ2−/− male (XY) and female (XX) medaka. b Sex-specific transcription of mTOR, HK2, ULK1, and Beclin1 (N = 12). c Liver estrogen synthesis (N = 10) in wild and ER−/− groups. d ER-dependent alteration of different autophagy genes (HK2, mTOR, NRF2, and LC3) validated by ChIP analysis (N = 5) at two independent sites. e Western blot panels of autophagy associated proteins among various groups. (N = 6, Number of times experiment was repeated). Tubulin was used as an internal control. Note: in graphs, data are plotted as means ± SEM; different alphabets denote significant differences at p < 0.05; NC negative control; tubulin-internal control. See also Fig. S3.
ERs directly controls LC3 transport
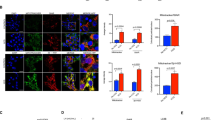
To investigate whether the reduction of autophagy at an earlier stage affected the late autophagic action, we measured the transcriptional activities of ULK1, Beclin1, and LC3, along with ULK1 phosphorylation, and quantified both the ULK1 and LC3 positive cell population. Significant diminution in the ULK1 and Beclin1 transcription and ULK1 phosphorylation was observed, while increased LC3 RNA amount was seen in the KO fish (Fig. 3b–e and S4A–C). This could suggest that the reduced ER amount affected the proper implementation of autophagy in the KO fish. To further validate the above findings, we performed cell sorting and found drastically diminished ULK1 positive cell population in both the KO groups, while the LC3 positive cells increased substantially in the ERα−/−-XX, followed by ERβ2−/−-XX fish (Fig. 4a and S4D). However, upon starvation, ULK1 and LC3 positive cells became abundant in both the sexes (Fig. S4E), thereby hinting that in order to cope an ER scarce condition, some alternative autophagic mechanism, which bypasses the canonical AMPK/ULK1 pathway, was induced [36]. Although we failed to observe any vacuole formation in the ER−/− livers, our immunoblotting analysis did reveal intensively aggregated LC3 expression in the hepatic nucleus of the ER−/− fish (ERα > ERβ2), while the control fish had dispersed LC3 expression in the cytoplasm (Fig. 4b). Despite starvation-associated alterations in the latter autophagic genes (Beclin1 and LC3), the autophagosome formation was presumably suppressed due to the mislocalization of LC3 protein (Figs. S4A-B and S5A) and subsequent interaction between LC3 and autophagic substrate like P62/SQSTM1 (Fig. 4c, d). Starvation has also been reported to increase the binding of SIRT1 with LC3 and activating the DOR protein in the nucleus, thus facilitating the LC3 nuclear-cytoplasm translocation [38]. So, we conducted another ChIP assay, and were able to determine the direct association between ERα-SIRT1, ERβ2-SIRT1, and ERα-DOR (Fig. 4e). The mRNA analysis also confirmed significant reduction of SIRT1 transcription in females of both KO groups, which could have resulted in the reduced acetylation of LC3 [39] (Fig. 4f and S5B). ERα−/−-specific downregulation of DOR transcription might have affected the LC3 translocation and finally reduced the autophagy in KO fish (Fig. 4f and S5B). Direct and specific interaction among ERs and different autophagy related genes are presumably the necessary checkpoints that need to be cleared/passed before a designated cell forms autophagosomes and brings about autophagy (Fig. S5C).
a FACS (Fluorescence activated cell sorting) of hepatic cell suspensions using ULK1 and LC3 antibody depicted sex-biased and ER−/− responsive alterations of ULK1/LC3 positive cells and increased LC3 positive ones and reduced ULK1 positive cells. b Immunoblot analysis of medaka primary hepatocyte cultures in wild, ERα−/− and ERβ2−/− hepatic cells. LC3 (green) and ULK1 (red) expression shows the differential nuclear-cytoplasmic localization of LC3. N = total occurrences of representative LC3 and ULK1 distribution/total number of cells observed. c, d Immunofluorescence confocal analysis of medaka primary hepatocyte cultures of wild, ERα−/−, and ERβ2−/− fish. c LC3 (green) and P62/SQSTM1 (white) expression among the various groups. The red box indicates the position of inset, and the inset (bounded in red box) shows the co-localization characteristics of LC3 and P62/SQSTM1. N = total occurrences of representative LC3 and P62/SQSTM1 distribution/total number of cells observed. d LC3 and P62/SQSTM1 co-localization puncta analysis in the cytoplasm and nucleus of wild, ERα−/−, and ERβ2−/− medaka primary hepatocyte. N = 15. e ChIP analysis of ERα and ERβ2 responsive SIRT1 and DOR alterations. f Real-time PCR profiling of SIRT1 and DOR gene in the wild and ER−/− groups. Note: in graphs, data are plotted as means ± SEM; different alphabets denote significant differences at p < 0.05; Scale = 10 μm. See also Fig. S4 and S5.
Discussion
Autophagy, regulated simultaneously in many tissues at the level of the whole organism, integrates both intracellular and endocrine signals, and is suggested to be regulated by autophagy flux, autophagy substrates, and steroid hormones [40, 41]. The huge signaling network downstream of estrogen promotes autophagy and also reduces overstimulated autophagy at the same time, which allows autophagy to be regulated by estrogen in a restricted range [5, 42]. The upregulation of steroid regulated autophagy is also associated with increased reactive oxygen species [43]. Our data demonstrate that starvation-induced oxidative stress responses are modulated by sex-biased estrogenic activity. Studies show that sexual biasness is highly tissue specific; for example, in rat, the autophagic responses are male biased in heart but female biased in kidney [44]. Such organ-specific difference can be attributed to the circulating steroids and/or difference in steroid receptor concentration [45]. In medaka, ERβ2 and ERα expressions are higher in female livers [35] and presently we depicted female biased liver autophagic response, thus highlighting the inter-correlation between steroid/steroid receptor and autophagy. This was further confirmed by E2 responsive alterations in HK2/mTOR concentration in the liver.
Upon starvation, E:T concentration changes and creates a situation where ER requires more transcription to cope up with the reduced ligand situation for maintaining estrogen/ER-associated physiological responses [46]. Multipoint direct ER-autophagic gene relations and starvation-related autophagy induction further suggests that ER concentrations are critical. Our data indicate that, in a single ER negative situation, compensatory response involving different ER was activated to maintain homeostasis and was further amplified upon starvation in the starved-ER−/− (compared with fed ER-KO) fish. In early phase of starvation, amino acid depletion regulates mTOR–ULK interaction but in later phase, when energy stores are depleted and lipid breakdown starts, the actions mostly depends on AMPK–ULK interaction [4]. Further investigation will decipher whether starvation-associated amplified response is a time-dependent phenomenon or not.
Nuclear hormone receptors are known to regulate the transcription of autophagy related genes [43]. Using ERα-KO or ERβ-KO mice, Kao et al. [13] found that ERα and ERβ orchestrate liver cell proliferation and differentiation, respectively, and acts as a quality control gate keeper for autophagy [47]. Recently we reported that, fish oocyte death can switch from apoptosis to autophagy based on concentration of steroid hormone [19] and ER knockdown induces autophagy in germ cells of medaka [18]. In the present study, we found that the constitutive autophagic responses and estrogen production were significantly reduced mostly in the ER−/− females. These facts suggest that threshold of both estrogen and ERs are important for autophagy. Our present data show that ERα inhibition lifted the negative regulation on NRF2/mTOR and probably executed excessive cell growth. Despite significantly increased mTOR transcription and ULK phosphorylation, LC3 production was induced in ERα−/− fish thereby confirming the existence of alternative autophagy. Similarly, it was shown that, inhibition of ERα signaling stimulates autophagosome formation and flux in ER+ breast cancer cells [48]. This might be a feedback mechanism to put a tight leash on autophagy by mTOR-regulated ER phosphorylation and anti-estrogen resistance [49]. However, mislocalization of LC3 in both ERα−/− and ERβ2−/− affected the final autolysosome formation and negated the effective execution of full-fledged alternative autophagy in ER−/− fish, and hence increased the P53 regulated apoptosis to control programmed cell death.
Cumulatively, our findings suggest that liver autophagy is influenced by the estrogen/ER concentration and exhibits female-biased response. The ER directed multipoint actions, especially LC3 nuclear-cytoplasmic transport is very critical for the autophagosomes formation and holds the rein to control excessive autophagic cell death. Since sex-biased disease management is gaining popularity and has tremendous potential for personalized gene therapy, this study is expected to improve our understanding about gender-specific remedial measures.
Materials and methods
Plasmid construction and mRNA transcription
pGEMT-easy plasmids carrying fragments of different genes were used for in situ hybridization (ISH) probe synthesis and qPCR standard preparation, wherever necessary. Expression plasmids were constructed, using pCS2(+) vector backbone and infusion cloning kit (Clonetech, USA), containing the complete ORFs of required genes, if not specifically mentioned. For overexpression (OV), HK2-5′-UTR, HK2 ORF-mCherry, and HK2 3′utr, and mCherry alone, were sequentially cloned into pCS2 vector, and named as pCS2-HK2-mCherry-OV, pCS2-mCherry-OV, respectively. The mRNA was transcribed with mMESSAGE mMACHINE T7 and SP6 kits (Ambion), following the manufacturer’s instructions. Poly A tail was added to the synthetic mRNA using Poly A tailing kit (Ambion).
Experimental animal, design, sampling, and statistical analysis
In this study, all animal treatments were conducted following the guidelines of Ehime University and Kyushu University, and were approved by the Institutional Animal Care and Use Committee of both Universities. Adult Japanese anchovy (Engraulis japonicas) and medaka (Oryzias latipes) cab strain were used in various experiments, if not otherwise mentioned. Japanese anchovy were maintained in flow-through water, at 24 ± 2 °C, 14 h light:10 h dark photoperiod, fed (3 times a day, @ 4% of their body weight) with commercially available pelleted feed (Otohime, Japan) [19]. Individual fish was biopsied to assess the gonadal maturity status and phenotypic sex and acclimatized in experimental tanks (100 fish/tank) for 2 weeks prior to the commencement of experiments. Medaka was maintained at 26 ± 2 °C, screened for genetic sex, using previously published protocols [18, 50], and divided into 30 fish/9 L tank, before commencing the experiments. Steroid doses and other conditions were determined using series of pilot experiments. Ten fish were sampled at each stage from each group. All in vivo experiments were conducted atleast thrice in duplicates, using three different mature populations of fish.
-
(1)
Starvation of Japanese anchovy: After the acclimation period, fish were subdivided into eight different tanks (stocking density of 100 fish/sex/3tonnes water). Fish in tanks 1–4 were fed with the commercial diet, while the fish in the remaining four tanks were starved continuously for a period of 20 days. Samplings were carried out at 2, 6, 12 and 20dat (days after treatment). The individual fish samples were treated as pseudo-replicate and tanks were used as replicate. Tank averages of each category were used for statistical analysis.
-
(2)
Estrogen and aromatase inhibitor (AI) exposure in Japanese anchovy: 17β-estradiol (E2, Wako, Japan) and AI (Exemestine, GmBH, Germany) were dissolved in ethanol, mixed with pelleted feed at two concentrations (10 and 100 mg/kg E2, and 10 and 100 mg/kg AI), dried overnight, fed to 100 male or female fish, respectively, for a period of 7 days and sampled. Vehicle controls, fed with ethanol soaked pelleted commercial feed, were similarly maintained and sampled. The experiments were repeated three times, and both design and analysis were similar to the abovementioned starvation experiment.
-
(3)
Starvation of medaka: Wild, ERα−/−, and ERβ2−/− homozygous medaka were used for this experiment. Each group of both genetic male and female medaka was subdivided into eight tanks. Half of them were kept in normal feeding regime and other half (remaining 4 tanks/group) were continuously starved for 7 days. Sampling was done at the end of experiment. The experiments were repeated three times, and both design and statistical analysis were similar to abovementioned starvation experiment.
-
(4)
Ex vivo cell culture: Japanese anchovy liver was cut into 5–6 mm sized cubes and randomly distributed on pre-arranged submersed cell-culture system. The submersed culture system was prepared by placing 3 cm2 nylon filter paper (Millipore) on a 1 cm3 agarose block kept in 1 ml of antibiotics (penicillin, streptomycine, neomycine, and gentamycin solution) containing L15 media (per well) in six-well plate. Three liver fragments were placed in each unit and incubated for 1 h, following which; extra 2 ml of antibiotic mixed L15 media was added. Each well was either treated with DMSO (vehicle control), 2 mM CaCl2, 10 nM CAI, 1 nM rapamycine, 1 nM nimphiridin, or 100 pM NSC. Twelve such plates, divided into four groups, i.e., SG+, S, G, SG−, were used in a single experiment. SG+, S, G wells were, respectively, supplemented with 10% FBS and 2 mM d-glucose, 10% FBS, and 2 mM d-glucose, while SG-wells contained only L15 medium, for a period of 24 h. All the incubations were performed at 27 °C. The concentration of each chemical was prefixed with pilot experiments. The experiment was repeated for 12 times, and average of each repeat was used for statistical analysis.
-
(5)
Ex vivo HK2 OV: Freshly excised livers from each sex were cut into 5–6 mm3 pieces and individually electroporated with CUY21 EDIT electroporator and CUY700P20 electrodes, following pre-adjusted electroporation protocol (16 v, 6 ms power on, 60 ms power off, 3 repeat). The tissue fragment was either electroporated with 10 µg/ml HK2-mcherry or mcherry mRNA, and respectively grouped as HK2-OV and mCherry. The electroporated tissue was immediately placed in chilled L15 medium for 15 min. Later all the liver fragments were similarly arranged and incubated in six-well type submerged culture units, and 8 plates/group/sex was prepared. Half of them were supplemented with 10% FBS and extra 2 mM glucose (SG+) while the other half were incubated in L15 medium only (SG−). Four hours post electroporation, either one of 2 m DMSO (vehicle control), 2 mM CaCl2, or 10 nM CAI was added to each well and cultured for 6 or 24 h at 27 °C before sampling. The experiment was repeated for 6 different time and average of each repeat was used for statistical analysis.
Sample collection
Tissue distribution samplings were carried out using mature adult fish (7/per sex). At each stage of sampling, parts of each liver were separately stored in RNAlater (Ambion, USA), or fixed using Bouin solution, wherever possible. After assessing the gonadal sexuality and maturity status (by assessing the abundance of sperm and vitellogenic oocytes) by histological analysis of Bouin-fixed gonads, ten individuals of each sex were randomly selected and used for RNA isolation. The embryos were grown at 24 °C, sampled at stage 39 (just before hatching) and stored in RNAlater for further analysis. For all experimental samples, parts of each experimental liver were snap-frozen in liquid nitrogen (for protein and enzymatic analysis), stored in PBS at −80 °C (for steroid analysis), stored in RNAlater (for Real-time PCR analysis), fixed in Bouin (for histology and immunohistochemistry (IHC)), or processed using PFA (for ISH, fluorescent ISH (FISH), FISH-fluorescent IHC (FIHC)). In case of medaka experiments, each experimental liver was stored, as a whole, in either PBS at −80 °C, RNAlater or fixed in Bouin, due to the small size. The ex vivo experimental samples from each unit were either stored in PBS at −80 °C, fixed in bouin or stored in RNAlater.
Mutant production and analysis
A gene-driven mutagenesis library (Kindly provided by NBRP medaka, Japan) was screened for ERα and ERβ2 mutation in the first exon of the ORF, following previously published protocols of HRM (high resolution analysis) [51, 52]. Any deviations from the wild DNA in melting curve were selected as mutant candidate. These selected DNAs were purified with ExoSAP-IT (GE healthcare, USA) and sequenced using a 3100-genetic analyzer (Life Technologies Corporation). The DNA carrying non-sense mutation was selected and in vitro fertilization of normal egg and the correspondent frozen sperm was performed to produce F0 progeny. The heterozygous genetic male and female F0 fish were separately backcrossed with wild Cab strains and two lines of progeny were produced. Similar crossing was performed repeatedly until F5 generation. Total 64 lines of F5 fish were histologically examined and the lines whose genetic and phenotypic sexuality are similar (XY fish with male phenotype and XX fish with female phenotype) were used for homozygous production (F6 generation) using sib mating. Although several lines were obtained, only two lines of ERα and ERβ2 mutants was used further analysis. F6 homozygous (−/−) and heterozygous (+/−) fish were used to produce maximum numbers of F7 homozygous fish. The present analysis was performed using F7, F8, and F9 generation of fish. Genotyping by tail clipping [50] was performed to ascertain sexuality and mutant status. The mutant analysis was further confirmed by sequencing.
cDNA cloning and sequence analysis
Total RNAs were extracted from medaka and Japanese anchovy liver, using direct-zol mini kit (Zymo, Japan) as previously described [53, 54]. One microgram of total RNA was reverse-transcribed into cDNA using the superscript IV (Invitrogen, Japan), following manufacturer’s instruction. Using publically available information at NCBI (http://www.ncbi.nlm.nih.gov/), degenerate primers for each gene was constructed and partial autophagy related genes were isolated and sequenced. RACE PCR was performed to isolate the complete cDNA sequence. In case of medaka, the ensemble medaka database (http://asia.ensembl.org/index.html) was used to identify the cDNA sequence followed by isolation and sequencing, using gene-specific primers. The putative amino acid sequences were translated using Expasy translation tools (http://web.expasy.org/translate) and phylogenetic analysis was performed using previously published protocols [54].
Quantification of changes in gene expression by real-time PCR
Changes in gene expression were quantified using the CFX-96 Real-time PCR system (Biorad, USA). Total RNA was isolated from cells, tissue fragment, liver, and other tissues using direct-zol mini kit (Zymo, Germany). cDNA synthesis was carried out using iScript™ Advanced cDNA Synthesis Kit (Biorad, USA) from 1 µg of total RNA. The first strand cDNAs were diluted to 200 µl for subsequent use. Gene-specific real-time PCR was performed using 2 µl of cDNA and SYBR green master mix (Applied Biosystem, USA), according to the manufacturer’s instructions. The PCR conditions included an initial denaturation at 94 °C (2 min) followed by 40 cycles at 94 °C (30 s) and 60 °C (1 min). Ef1α and β-actin were used as internal controls. The absolute transcript copy number of each gene was determined with the help of appropriate standard curves and normalized with the geometric mean Ef1α and β-actin copy numbers in each sample. Average relative expression derived from triplicates was used for further analysis. The specificity of primer sets, throughout this range of detection, was confirmed by the observation of a single amplification product of the expected size, Tm (melting temperature) and sequences. All assays were quantified, with standard curves (mean Ct vs log cDNA dilution) having slopes between −2.99 and −3.34, a linear correlation (R2) between the mean Ct and the logarithm of cDNA dilution of >0.985 in each case. All test cDNAs were run in duplicates for each gene.
Histology, ISH, FISH, and FIHC
Bouin fixed, paraffin-embedded samples (at least 10 fish per group) were used for standard Hematoxylin & Eosin (HE) staining and FIHC, while, 4% PFA fixed samples were used for ISH and FISH. All the histological analyses were performed using 5-µm sections. For ISH, sense and anti-sense fluorescein or digoxigenin-labeled RNA probes were transcribed in vitro, using RNA labeling kit (Roche Diagnostics GmbH, Mannheim, Germany), from plasmid DNA containing the respective genes. Sections were deparaffinised, hydrated, treated with proteinase K at 10 µg/ml (Roche), and hybridized with the sense or anti-sense labeled RNA probes at 60 °C for 18–24 h. The hybridization signals were detected following previously published protocols [54].
The FIHC procedures are same as previously described [55]. Briefly, 5 µm bouin-fixed tissue sections were treated for antigen retrieval, peroxidase activity and blocked in 2% FBS-PBS, incubated with primary (1:500 dilution) and Alexa-tagged secondary (1:1000 dilution) antibody, and micrographed using a confocal microscope (Axio 710, Zeiss, Germany). HK2, mTOR, AMPK, ULK1, Beclin1, LC3, P53, ERα, and ERβ antibodies were used in our study.
Transmission electron microscopy
Fresh liver fragments of 1–2 mm3 size were immediately fixed in cold 2% paraformaldehyde (PFA) and 2% glutaraldehyde (GA) in 0.1 M phosphate buffer (PB) pH 7.4 at 4 °C overnight. On the following day, the samples were washed with 0.1 M PB 3 times for 30 min each, post-fixed with 2% osmium tetroxide in 0.1 M PB for 2 h, dehydrated in graded ethanol solutions (50, 70, 90, and 100%), infiltrated with propylene oxide (PO) two times for 20 min each, put into a 70:30 mixture of PO and resin (Quetol-812; Nisshin EM co., Tokyo, Japan) for 1 h and then the PO was volatilized by keeping the tube cap open overnight. The samples were transferred to a fresh 100% resin and were polymerized at 60 °C for 48 h. The polymerized resins were ultra-thin sectioned at 70 mm with a diamond knife using an ultramicrotime (Ultracut UCT; Leica, Vienna, Austria), mounted on copper grids, stained with 2% uranyl acetate at room temperature for 15 min, and then washed with distilled water, and was further secondary-stained with lead stain solution (Sigma-Aldrich Co., Tokyo, Japan) at room temperature for 3 min. The grids were observed by a transmission electron microscope (JEM-1400Plus; JEOL Ltd., Tokyo, Japan) at an acceleration voltage of 100 kV. Digital images (3296 X 2472 pixels) were taken with a CCD camera (EM-14830RUBY2; JEOL Ltd., Tokyo, Japan).
Western blotting
The concentrations of crude liver protein extracts were determined by the Thermo Scientific Pierce BCA Protein Assay Kit and uniformized to 20 mg/ml, 15 µl of each sample was loaded and separated by SDS-PAGE, and then transferred onto a polyvinylidene fluoride (PVDF) membrane (Merck Millipore, German). The membranes were incubated with diluted antibody (1:10000) overnight, and then with 1:20,000 dilution of secondary goat anti-rabbit/mouse HRP-conjugated Ab (Vector Laboratories) for 1 h. The membranes were washed with TBS-tween 20 (20 mM tris-base, 150 mM NaCl, 0.05% tween 20, pH 7.5) for several times after each incubation. PVDF blocking buffer (Toyobo, Japan) along with respective IgG was used for effective blocking, while the antibodies were diluted with Can get signal (Toyobo, Japan). Finally, band revelations were achieved by Pierce ECL Plus Western Blot Kit (Thermofisher Scientific, Japan) and imaging was done using ImageQuant LAS-4000 (GE healthcare, USA). Hexokinase (GTX124375), mTOR (GTX124771), P62 (GTX100685), LC3 (GTX127375 and GTX82986), Beclin (GTX31722), ULK1 (GTX80551), P53 (GTX128135) GAPDH (GTX100118) and Tubulin (GTX6288002) antibodies were procured from Genetex, Japan. Phospho ULK1 S757 (#6888) was purchased from Cell signaling.
Steroid measurement
Quick-frozen individual liver samples were thawed, weighed, homogenized, and used for steroid extraction using diethyl ether. Serum samples were similarly used for steroid extraction whenever necessary. The freeze-dried extracts were diluted in appropriated quantity of dilution buffer (@ 20 µl per mg tissue) and used for testosterone measurement, using testosterone high sensitivity ELISA kit (Enzo, Japan), following manufacturer’s instructions. The samples were further diluted (100 times) to measure the estrogen, using 17β-estradiol high sensitivity ELISA kit (Enzo, Japan). The steroid concentrations were calculated based on the standard curve prepared using respective steroids provided by the manufacturer (Enzo, Japan). Both tissue and extract duplicates were analyzed before final calculation. Preliminary analysis was carried out to standardize the sampling method and assessment protocols.
Autophagosome estimation
Fluorescence activate cell sorting and live cell immunoblotting
In total, 1 mm3 blood cell free liver fragment was biopsied and incubated in L15 media containing 0.25% Trypsin for 30 min, with periodical shaking, to prepare a single cell suspension. The enzymatic digestion was stopped using 2% FBS and the cell suspension was immediately fixed with 2% formaldehyde-PBS for 15 min at 4 °C, washed three times with PBS and re-suspended in blocking solution (PBS containing 2% FBS and 0.001% Triton X). One hour after incubation the cells were pelleted, incubated with Alexa-tagged antibodies for 1 h, washed several times with PBST (PBS + 0.01% tween 20), and subjected to cell sorting using Shimadzu PER-FLOW (Tokyo, Japan) cell sorter, following pre-adjusted protocols of hepatic cell sorting.
The remaining liver were similarly trypsinized to produce single cell suspension and centrifuged at low-speed centrifugation at gradually increasing rates (30 × g, 40 × g, and 50 × g, for 10 min) to pellet the primary hepatocytes. The pellets were then washed with complete L15 medium and incubated in complete L15 medium for 30 min. Later the cells pellets were incubated with mouse-ASGR1 antibody (Thermofisher Scientific, Japan, MA1-40244) followed by magnetic bead sorting using pre-blocked anti-mouse IgG dyna beads to obtain a pure population of hepatocytes. Finally, the bead-cell suspensions were treated with TrypLE (Thermofisher Scientific, Japan) for 5 min, removed beads using a magnetic stand, and hepatocyte suspension were collected using low-speed centrifugation at 100 × g for 10 min. A total of 105 cells/100 µl complete L15 medium were plated on attachment factor (Invitrogen, USA) coated plastic cover slip mounted on each wells of a 24-well plate, incubated for 6 h at 33 °C, then 1 ml of L15 complete medium was added, and further incubated for 42 h, with daily medium exchange for complete attachment and 60–70% confluency. Two days after incubation, the cover slips were carefully washed with L15 medium and divided into two groups, namely, Starved and Fed, respectively, incubated in 20 times diluted L15 medium and complete L15 medium, for 4 h. Bafilomycin A1 treatments were initiated 2 h before fixation, wherever necessary. The cells were fixed using 2% formaldehyde for 20 min, perforated with glycine, washed with PBS, blocked using 2% FBS-PBS-0.001% Triton X for 1 h, incubated with primary (1:300) antibody overnight, washed and re-incubated with fluorescent tagged secondary antibody (1:1000) overnight, and finally mounted on glass slides for microscopic observation and photomicrographed using an Axio-710 confocal microscope (Zeiss). The images were quantified using the default ‘Analyze particles’ plugin in ImageJ to analyze the hepatocyte LC3 puncta.
Chromatin immunoprecipitation (ChIP)
Atleast 25 kb upstream sequence (from the ORF start site) were obtained from ensemble (http://asia.ensembl.org/index.html), and the promoter regions were identified using promoter 2.0 prediction server (http://www.cbs.dtu.dk/services/Promoter/). Ten kilobyte genomic region, including the promoter region, was further analyzed with Dragon ERE finder version 3.0 (http://datam.i2r.a-star.edu.sg/ereV3/), and two most potential ERE sites were predicted. For in vivo ChIP analysis, the ChIP expression-shearing kit (Active motif) was used according to the manufacturer’s instructions, using 20 mg of XX or XY liver samples (ten fish each), ERα and ERβ antibody (3 mg, Active motif, 61035 and 39767, respectively). Preliminary experiments were conducted with 20 mg of liver samples from ERα-GFP or ERβ2-GFP mRNA-injected fish (10 adults, injected along with TransIT®-QR Delivery Solution (Mirus-bio, USA)) and GFP monoclonal antibody (3 mg, AbCAM, ab290), to validate the ER specificity. The ERα- and ERβ2-GFP plasmids were constructed by sequentially fusing ERα-/β2-5′UTR, ERα-/β2- ORF (stop codon less), eGFP, and ERα-/β2-3′UTR into a pCS2 vector, using infusing cloning kit (Clonetech). RNA was synthesized with mMESSAGE mMECHINE SP6 kit (Ambion, USA) following poly A addition using Poly A tailing kit (Ambion). After tissue disaggregation and cell re-suspension, DNA was sheared, according to the manufacturer’s protocols. The ChIP procedure using ERα-/β2-GFP mRNA was validated as described earlier [50].
Data analysis
Statistical differences in relative mRNA expression between various experimental groups were assessed by One or two-way ANOVA of normalized data, followed by Tukey’s test or Student’s t test. All statistical analyses were performed using SPSS, version 22. All experimental data are shown as mean ± SEM. Differences were considered statistically significant at p < 0.05, if not otherwise mentioned. The correlations were calculated using the Pearson correlation coefficient method.
References
Lindqvist LM, Simon AK, Baehrecke EH. Current questions and possible controversies in autophagy. Cell Death Discov. 2015;1:15036.
Ezaki J, Matsumoto N, Takeda-Ezaki M, Komatsu M, Takahashi K, Hiraoka Y, et al. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy. 2011;7:727–36.
Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75.
Madrigal-Matute J, Cuervo AM. Reviews in basic and clinical gastroenterology and hepatology. Gastroenterology. 2016;150:328–39.
Zhu W, Qu H, Xu K, Jia B, Li H, Du Y, et al. Differences in the starvation-induced autophagy response in MDA-MB-231 and MCF-7 breast cancer cells. Anim Cells Syst. 2017;21:190–8.
Komatsu M, Ueno T, Waguri S, Uchiyama Y, Kominami E, Tanaka K. Constitutive autophagy: vital role in clearance of unfavorable proteins in neurons. Cell Death Differ. 2007;14:887–94.
Rautou PE, Mansouri A, Lebrec D, Durand F, Valla D, Moreau R. Autophagy in liver diseases. J Hepatol. 2010;53:1123–34.
Lista P, Straface E, Brunelleschi S, Franconi F, Malorni W. On the role of autophagy in human diseases: a gender perspective. J Cell Mol Med. 2011;15:1443–57.
Shen M, Shi H. Sex hormones and their receptors regulate liver energy homeostasis. Int J Endocrinol. 2015;2015:294278.
Totta P, Busonero C, Leone S, Marino M, Acconcia F. Dynamin II is required for 17β-estradiol signaling and autophagy-based ERα degradation. Sci Res. 2016;6:23727.
Xiang J, Liu X, Ren J, Chen K, Wang H-I, Miao YY, et al. How does estrogen work on autophagy? Autophagy. 2019;15:197–211.
Gustafsson JA. What pharmacologists can learn from recent advances in estrogen signalling. Trends Pharm Sci. 2003;24:479–85.
Kao T-L, Kuan Y-P, Cheng W-C, Chang W-C, Jeng L-B, Yeh S, et al. Estrogen receptors orchestrate cell growth and differentiation to facilitate liver regeneration. Theranostics. 2018;8:2672–82.
Durazzo M, Belci P, Collo A, Prandi V, Pistone E, Martorana M, et al. Gender specific medicine in liver diseases: a point of view. World J Gastroenterol. 2014;20:2127–35.
Schartl M. Beyond the zebrafish: diverse fish species for modelling human diseases. Dis Model Mech. 2014;7:181–92.
Yabu T, Imamura S, Mizusawa N, Touhata K, Yamashita M. Induction of autophagy by amino acid starvation in fish cells. Mar Biotechnol. 2012;14:491–501.
Varga M, Sass M, Papp D, Takacs-Vellai K, Kobolak J, Dinnyes A, et al. Autophagy is required for zebrafish caudal fin regeneration. Cell Death Differ. 2014;21:547–56.
Chakraborty T, Mohapatra S, Zhou LY, Matsubara T, Iguchi T, Nagahama Y. Estrogen receptor β2 oversees germ cell maintenance and gonadal sex differentiation in medaka, Oryzias latipes. Stem Cell Rep. 2019;13:419–33.
Chakraborty T, Mohapatra S, Tobayama M, Ohta K, Ryu YW, Kazeto Y, et al. Hatching enzymes disrupt aberrant gonadal degeneration by the autophagy/apoptosis cell fate decision. Sci Rep. 2017;7:3183.
Sakaguchi K, Yoneda M, Sakai N, Nakashima K, Kitano H, Matsuyama M. Comprehensive experimental system for a promising model organism candidate for marine teleosts. Sci Rep. 2019;9:4948.
Qiao Q, Le Manach S, Sotton B, Huet H, Duvernois-Berthet E, Paris A, et al. Deep sexual dimorphism in adult medaka fish liver highlighted by multi-omic approach. Sci Rep. 2016;6:32459.
Antonioli M, Albiero F, Fimia GM, Piacentini M. AMBRA1-regulated autophagy in vertebrate development. Int J Dev Biol. 2015;59:109–17.
Shi H, Seeley RJ, Clegg DJ. Sexual differences in the control of energy homeostasis. Front Neuroendocrinol. 2009;30:396–404.
Wagner EJ. Sex differences in cannabinoid-regulated biology: a focus on energy homeostasis. Front Neuroendocrinol. 2016;40:101–9.
Brunelli E, Domanico F, La Russa D, Pellegrino D. Sex differences in oxidative stress biomarkers. Curr Drug Targets. 2014;15:811–5.
Zielniok K, Motyl T, Gajewska M. Functional interactions between 17β-Estradiol and progesterone regulate autophagy during acini formation by bovine mammary epithelial cells in 3D cultures. Biomed Res Int. 2014;2014:382653.
Hibshman JD, Leuthner TC, Shoben C, Mello DF, Sherwood DR, Meyer JN, et al. Nonselective autophagy reduces mitochondrial content during starvation in Caenorhabditis elegans. Am J Physiol Cell Physiol. 2018;315:C781–C792.
Baranowski Z, Hrebenda B, Cieslawska M, Beylina SI. Division of Phusarum mitochondria during starvation. Cell Biol Int Rep. 1991;15:197–204.
Chen JQ, Yager JD. Estrogen’s effects on mitochondrial gene expression: Mechanisms and potential contributions to estrogen carcinogenesis. Ann NY Acad Sci. 2004;1028:258–72.
Chen JQ, Yager JD, Russo J. Regulation of mitochondrial respiratory chain structure and function by estrogens/estrogen receptors and potential physiological/pathophysiological implications. Biochim Biophys Acta. 2005;1746:1–17.
Guevara R, Gianotti M, Roca P, Oliver J. Age and sex-related changes in rat brain mitochondrial function. Cell Physiol Biochem. 2011;27:201–6.
Filomeni G, De Zio D, Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2015;22:377–88.
Dai W, Li Y, Zheng H. Estradiol/Testosterone imbalance: impact on coronary heart disease risk factors in post-menopausal women. Cardiology. 2012;121:249–54.
Campesi I, Straface E, Occhioni S, Montella A, Franconi F. Protein oxidation seems to be linked to constitutive autophagy: a sex study. Life Sci. 2013;93:145–52.
Chakraborty T, Shibata Y, Zhou LY, Katsu Y, Iguchi T, Nagahama Y. Differential expression of three estrogen receptor subtype mRNAs in gonads and liver from embryos to adults of the medaka, Oryzias latipes. Mol Cell Endocrinol. 2011;333:47–54.
Grotemeier A, Alers S, Pfisterer SG, Paasch F, Daubrawa M, Dieterle A, et al. AMPK-independent induction of autophagy by cytosolic Ca2+ increase. Cell Signal. 2010;22:914–25.
Goudarzi KM, Nister M, Lindstrom MS. mTOR inhibitors blunt the p53 response to nucleolar stress by regulating RPL11 and MDM2 levels. Cancer Biol Ther. 2014;15:1499–514.
Huang R, Xu Y, Wan W, Shou X, Qian J, You Z, et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell. 2015;57:456–66.
Li X, Wang Y, Xiong Y, Wu J, Ding H, Chen X, et al. Galangin induces autophagy via deacetylation of LC3 by SIRT1 in HEPG2 cells. Sci Rep. 2016;6:30496.
Klionsky DJ, Abdelmohsen L, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12:1–222.
Sinha RA, Singh BK, Yen PM. Reciprocal crosstalk between autophagy and endocrine signaling in metabolic homeostasis. Endocrin Rev. 2017;38:69–102.
Petherick KJ, Williams AC, Lane JD, Ordóñez-Morán P, Huelsken J, Collard TJ, et al. Autolysosomal β-catenin degradation regulates Wnt-autophagy-p62 crosstalk. EMBO J. 2013;32:1903–16.
Park JM, Jung CH, Seo M, Otto NM, Grunwald D, Kim KH, et al. The ULK1 complex mediates MTORC1 signaling to the autophagy initiation machinery via binding and phosphorylating ATG14. Autophagy. 2016;12:547–64.
Campesi I, Straface E, Occhioni S, Fenu G, Franconi F, Montella A. Sex differences in redox state, autophagy and lysosomal function. Ital J Anat Embryol. 2013;118:43.
Campesi I, Occhioni S, Fois M, Capobianco G, Dessole S, Franconi F, et al. Autophagic process is sexually different in VSMCs from human male and female neonates. Ital J Anat Embryol. 2015;120:91.
Słuczanowska-Głąbowska S, Laszczyńska M, Piotrowska K, Grabowska M, Grymuła K, Ratajczak MZ. Caloric restriction increases ratio of estrogen to androgen receptors expression in murine ovaries–potential therapeutic implications. J Ovarian Res. 2015;8:57.
Wei Y, Huang C, Wu H, Huang J. Estrogen receptor beta (ERβ) mediated-cyclinD1 degradation via autophagy plays an anti-proliferation role in colon cells. Int J Biol Sci. 2019;15:942–52.
Cook KL, Clarke R. Estrogen receptor-α signaling and localization regulates autophagy and unfolded protein response activation in ER+ breast cancer. Receptors Clin Investig. 2014;1:e316.
Yamnik RL, Holz MK. mTOR/S6K1 and MAPK/RSK signaling pathways coordinately regulate estrogen receptor α serine 167 phosphorylation. FEBS Lett. 2010;584:124–8.
Chakraborty T, Zhou LY, Chaudhari A, Iguchi T, Nagahama Y. Dmy initiates masculinity by altering Gsdf/Sox9a2/Rspo1 expression in medaka (Oryzias latipes). Sci Rep. 2016;6:19480.
Taniguchi Y, Takeda S, Furutani-Seiki M, Kamei Y, Todo T, Sasado T, et al. Generation of medaka gene knockout models by target-selected mutagenesis. Genome Biol. 2006;7:R116.
Ishikawa T, Kamei Y, Otozai S, Kim J, Sato A, Kuwahara Y, et al. High-resolution melting curve analysis for rapid detection of mutations in a Medaka TILLING library. BMC Mol Biol. 2010;11:70.
Mohapatra S, Chakraborty T, Reza MAN, Shimizu S, Matsubara T, Ohta K. Short-term starvation and realimentation helps stave off Edwardsiella tarda infection in red sea bream (Pagrus major). Comp Biochem Physiol B. 2017;206:42–53.
Mohapatra S, Chakraborty T, Miyagawa S, Zhou LY, Ohta K, Iguchi T, et al. Steriod responsive regulation of IFNγ2 alternative splicing and its possible role in germ cell proliferation in medaka. Mol Cell Endocrinol. 2015;400:61–70.
Zhou LY, Chakraborty T, Zhou Q, Mohapatra S, Nagahama Y, Zhang YG. Rspo-1 activated signalling molecules are sufficient to induce ovarian differentiation in XY medaka (Oryzias latipes). Sci Rep. 2016;6:19543.
Acknowledgements
We thank the National Bioresource Project Medaka for supplying the various medaka strains. The authors are grateful to Yukiko Ogino (Associate Professor, Kyushu University, Japan) and Yukinori Kazeto (Senior Scientist, National fisheries research agency, Japan) for their critical reading and valuable constructive review of the manuscript. This work was in part supported by Grants from Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan; Japanese Society for the Promotion of Science (JSPS) Kakenhi, Grant Nos. 23688022, 24370028, 16H04981, 23380110, 18K14520, and 19H03049; Sumitomo Grant no. 180959; and Ehime University Post-Doctoral Research Grant, Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by H. Zhang
Rights and permissions
About this article
Cite this article
Mohapatra, S., Chakraborty, T., Shimizu, S. et al. Estrogen and estrogen receptors chauffeur the sex-biased autophagic action in liver. Cell Death Differ 27, 3117–3130 (2020). https://doi.org/10.1038/s41418-020-0567-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-020-0567-3
This article is cited by
-
Estrogen inhibits endoplasmic reticulum stress and ameliorates myocardial ischemia/reperfusion injury in rats by upregulating SERCA2a
Cell Communication and Signaling (2022)
-
Introducing ADNP and SIRT1 as new partners regulating microtubules and histone methylation
Molecular Psychiatry (2021)