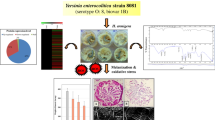
Abstract
Objective
Earlier, we have found that the enteropathogenic Yersinia enterocolitica have evolved the survival mechanisms that regulate the expression of laccase-encoding genes in the gut. The present study aims to characterize the purified recombinant laccase from Y. enterocolitica strain 8081 biovar 1B and understand its effect on the midgut of cotton bollworm, Helicoverpa armigera (Hübner) larvae.
Results
The recombinant laccase protein showed high purity fold and low molecular mass (~ 43 kDa). H. armigera larvae fed with laccase protein showed a significant decrease in body weight and damage in the midgut. Further, transmission electron microscopy (TEM) studies revealed the negative effect of laccase protein on trachea, malpighian tubules, and villi of the insect. The proteome comparison between control and laccase-fed larvae of cotton bollworm showed significant expression of proteolytic enzymes, oxidoreductases, cytoskeletal proteins, ribosomal proteins; and proteins for citrate (TCA cycle) cycle, glycolysis, stress response, cell redox homeostasis, xenobiotic degradation, and insect defence. Moreover, it also resulted in the reduction of antioxidants, increased melanization (insect innate immune response), and enhanced free radical generation.
Conclusions
All these data collectively suggest that H. armigera (Hübner) larvae can be used to study the effect of microbes and their metabolites on the host physiology, anatomy, and survival.
Graphic abstract









Similar content being viewed by others
References
Achard ME, Tree JJ, Holden JA, Simpfendorfer KR, Wijburg OL, Strugnell RA, Schembri MA, Sweet MJ, Jennings MP, McEwan AG (2010) The multi-copper-ion oxidase CueO of Salmonella enterica serovar Typhimurium is required for systemic virulence. Infect Immun 78:2312–2319. https://doi.org/10.1128/IAI.01208-09
Ahlawat S, Singh D, Virdi JS, Sharma KK (2019) Molecular modeling and MD-simulation studies: fast and reliable tool to study the role of low-redox bacterial laccases in the decolorization of various commercial dyes. Environ Pollut 253:1056–1065. https://doi.org/10.1016/j.envpol.2019.07.083
Akbar SM, Sharma HC, Jayalakshmi SK, Sreeramulu K (2012) Effect of pyrethroids, permethrin and fenvalarate, on the oxidative stress of Helicoverpa armigera. World J Sci Technol 2:1–5
Assar AA, El-Mahasen MA, Dahi HF, Amin HS (2016) Biochemical effects of some insect growth regulators and bioinsecticides against cotton leafworm, Spodoptera littoralis (Boisd.) (Lepidoptera: Noctuidae). J Biosci Appl Res 8:582–589
Ayobola A (2012) Assessment of lipid peroxidation and activities of antioxidant enzymes in phosphide-powder residue exposed rats. J Drug Metab Toxicol 3(5):132. https://doi.org/10.4172/2157-7609.1000132
Barison N, Gupta R, Kolbe M (2013) A sophisticated multi-step secretion mechanism: how the type 3 secretion system is regulated. Cell Microbiol 15(11):1809–1817. https://doi.org/10.1111/cmi.12178
Becker KW, Skaar EP (2014) Metal limitation and toxicity at the interface between host and pathogen. FEMS Microbiol Rev 38:1235–1249. https://doi.org/10.1111/1574-6976.12087
Beers R, Sizer IW (1952) A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem 195:133
Bresolin G, Morgan JA, Ilgen D, Scherer S, Fuchs TM (2006) Low temperature-induced insecticidal activity of Yersinia enterocolitica. Mol Microbiol 59(2):503–512. https://doi.org/10.1111/j.1365-2958.2005.04916.x
Burnens AP, Frey A, Nicolet J (1996) Association between clinical presentation, biogroups and virulence attributes of Yersinia enterocolitica strains in human diarrhoeal disease. Epidemiol Infect 116:27–34
Büyükgüzel E, Büyükgüzel K, Snela M, Erdem M, Radtke K, Ziemnicki K, Adamski Z (2013) Effect of boric acid on antioxidant enzyme activity, lipid peroxidation, and ultrastructure of midgut and fat body of Galleria mellonella. Cell Biol Toxicol 29(2):117–129. https://doi.org/10.1007/s10565-013-9240-7
Cerenius L, Söderhäll K (2012) Crustacean immune responses and their implications for disease control. Infectious disease in aquaculture. Woodhead Publishing, Sawston, pp 69–87
Chen B, Zhang D, Wang X, Ma W, Deng S, Zhang P et al (2017) Proteomics progresses in microbial physiology and clinical antimicrobial therapy. Eur J Clin Microbiol Infect Dis. https://doi.org/10.1007/s10096-016-2816-4
de Groot NS, Ventura S (2006) Effect of temperature on protein quality in bacterial inclusion bodies. FEBS Lett 580(27):6471–6476. https://doi.org/10.1016/j.febslet.2006.10.071
Demaeght PA (2015) A genomic approach to investigate resistance mechanisms in the two-spotted spider mite Tetranychus urticae. 9789491407161.
Dhar MS, Virdi JS (2013) Interaction of Yersinia enterocolitica biovar 1A with cultured cells in vitro does not reflect the two previously identified clonal groups. J Med Microbiol 62:1807–1814. https://doi.org/10.1099/jmm.0.056077-0
Fallman M, Deleuil F, McGee K (2001) Resistance to phagocytosis by Yersinia. Int J Med Microbiol 291(6–7):501–509. https://doi.org/10.1078/1438-4221-00159
Fang W, Fernandes ÉK, Roberts DW, Bidochka MJ, Leger RJS (2010) A laccase exclusively expressed by Metarhizium anisopliae during isotropic growth is involved in pigmentation, tolerance to abiotic stresses and virulence. Fungal Genet Biol 47(7):602–607. https://doi.org/10.1016/j.fgb.2010.03.011
Fernandes AT, Soares CM, Pereira MM, Huber R, Grass G, Martins LO (2007) A robust metallo-oxidase from the hyperthermophilic bacterium Aquifex aeolicus. FEBS J 274(11):2683–2694. https://doi.org/10.1111/j.1742-4658.2007.05803.x
Fernández-Sánchez C, Tzanov T, Gübitz GM, Cavaco-Paulo A (2002) Voltammetric monitoring of laccase-catalysed mediated reactions. Bioelectrochemistry 58:149–156. https://doi.org/10.1016/S1567-5394(02)00119-6
Greenwood MH, Hooper WL (1990) Excretion of Yersinia spp. associated with consumption of pasteurized milk. Epidemiol Infect 104:345–350. https://doi.org/10.1017/S0950268800047361
Gutteridge JM (1986) Iron promoters of the Fenton reaction and lipid peroxidation can be released from haemoglobin by peroxides. FEBS Lett 201(2):291–295
Hall SJ, Hitchcock A, Butler CS, Kelly DJ (2008) A multicopper oxidase (Cj1516) and a CopA homologue (Cj1161) are major components of the copper homeostasis system of Campylobacter jejuni. J Bacteriol 190:8075–8085. https://doi.org/10.1128/JB.00821-08
Haque MR, Higashiura A, Nakagawa A, Hirowatari A, Furuya S, Yamamoto K (2019) Molecular structure of a 5, 10-methylenetetrahydrofolate dehydrogenase from the silkworm Bombyx mori. FEBS Open Bio 9(4):618–628. https://doi.org/10.1002/2211-5463.12595
Heermann R, Fuchs TM (2008) Comparative analysis of the Photorhabdus luminescens and the Yersinia enterocolitica genomes: uncovering candidate genes involved in insect pathogenicity. BMC Genomics 9(1):40. https://doi.org/10.1186/1471-2164-9-40
Howard SL, Gaunt MW, Hinds J, Witney AA, Stabler R, Wren BW (2006) Application of comparative phylogenomics to study the evolution of Yersinia enterocolitica and to identify genetic differences relating to pathogenicity. J Bacteriol 188:3645–3653. https://doi.org/10.1128/JB.188.10.3645-3653.2006
Hu X, Chen L, Xiang X, Yang R, Yu S, Wu X (2012) Proteomic analysis of peritrophic membrane (PM) from the midgut of fifth-instar larvae, Bombyx mori. Mol Biol Rep 39(4):3427–3434. https://doi.org/10.1007/s11033-011-1114-6
Hunke S, Betton JM (2003) Temperature effect on inclusion body formation and stress response in the periplasm of Escherichia coli. Mol Microbiol 50(5):1579–1589. https://doi.org/10.1046/j.1365-2958.2003.03785.x
Ighodaro OM, Akinloye OA (2018) First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): their fundamental role in the entire antioxidant defence grid. Alex Med J 54(4):287–293. https://doi.org/10.1016/j.ajme.2017.09.001
Jain KK, Kumar A, Shankar A, Pandey D, Chaudhary B, Sharma KK (2019) De novo transcriptome assembly and protein profiling of copper-induced lignocellulolytic fungus Ganoderma lucidum MDU-7 reveals genes involved in lignocellulose degradation and terpenoid biosynthetic pathways. Genomics. https://doi.org/10.1016/j.ygeno.2019.01.012
Jia FX, Dou W, Hu F, Wang JJ (2011) Effects of thermal stress on lipid peroxidation and antioxidant enzyme activities of oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae). Fla Entomol 94(4):956–964
Kamal AHM, Fessler MB, Chowdhury SM (2018) Comparative and network-based proteomic analysis of low dose ethanol- and lipopolysaccharide-induced macrophages. PLoS ONE 13(2):e0193104. https://doi.org/10.1371/journal.pone.0193104
Kamen DE, Woody RW (2002) Folding kinetics of the protein pectate lyase C reveal fast forming intermediate and slow proline isomerisation. Biochemistry 41:4713–4723. https://doi.org/10.1021/bi0115129
Kim C, Lorenz WW, Hoopes JT, Dean JF (2001) Oxidation of phenolate siderophores by the multicopper oxidase encoded by the Escherichia coli yacK gene. J Bacteriol 183:4866–4875. https://doi.org/10.1128/JB.183.16.4866-4875.2001
Kumar A, Sharma KK, Kumar P, Ramchiary N (2015) Laccase isozymes from Ganoderma lucidum MDU-7: isolation, characterization, catalytic properties and differential role during oxidative stress. J Mol Catal B 113:68–75. https://doi.org/10.1016/j.molcatb.2015.01.010
Kumar V, Bal A, Gill KD (2008) Impairment of mitochondrial energy metabolism in different regions of rat brain following chronic exposure to aluminium. Brain Res 232:94–103. https://doi.org/10.1016/j.brainres.2008.07.028
Leclercq A, Martin L, Vergnes ML, Ounnoughene N, Laran JF, Giraud P, Carniel E (2005) Fatal Yersinia enterocolitica biotype 4 serovar O: 3 sepsis after red blood cell transfusion. Transfusion 45:814–818. https://doi.org/10.1111/j.1537-2995.2005.04363.x
Ley C, Çekiҫ SZ, Kochius S, Mangoldb KM, Schwaneberg U, Schrader J, Holtmann D (2013) An electrochemical microtiter plate for parallel spectroelectrochemical measurements. Electrochim Acta 89:98–105. https://doi.org/10.1016/j.electacta.2012.10.151
Li S, Yu X, Feng Q (2019) Fat body biology in the last decade. Annu Rev Entomol 64:315–333
Liu G, Ma H, Xie H, Xuan N, Guo X, Fan Z et al (2016) Biotype characterization, developmental profiling, insecticide response and binding property of Bemisia tabaci chemosensory proteins: role of CSP in insect defense. PLoS ONE 11(5):e0154706. https://doi.org/10.1371/journal.pone.0154706
Lu A, Zhang Q, Zhang J, Yang B, Wu K, Xie W et al (2014) Insect prophenoloxidase: the view beyond immunity. Front Physiol 5:252. https://doi.org/10.3389/fphys.2014.00252
Ma S, Liu N, Jia H, Dai D, Zang J, Cao Z, Dong J (2018) Expression, purification, and characterization of a novel laccase from Setosphaeria turcica in Eschericha coli. J Basic Microbiol 58(1):68–75. https://doi.org/10.1002/jobm.201700212
Martins LO, Soares CM, Pereira MM, Teixeira M, Costa T, Jones GH, Henriques AO (2002) Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural component of the Bacillus subtilis endospore coat. J Biol Chem 277:18849–18859. https://doi.org/10.1074/jbc.M200827200
Mollania N, Khajeh K, Ranjbar B, Rashno F, Akbari N, Fathi-Roudsari M (2013) An efficient in vitro refolding of recombinant bacterial laccase in Escherichia coli. Enzym Microb Technol 52(6–7):325–330. https://doi.org/10.1016/j.enzmictec.2013.03.006
Mustacich D, Powis G (2000) Thioredoxin reductase. Biochem J 346(1):1–8
Palmer LD, Skaar EP (2016) Transition metals and virulence in bacteria. Annu Rev Genet 50:67–91. https://doi.org/10.1146/annurev-genet-120215-035146
Papaneophytou CP, Kontopidis G (2014) Statistical approaches to maximize recombinant protein expression in Escherichia coli: a general review. Protein Expr Purif 94:22–32. https://doi.org/10.1016/j.pep.2013.10.016
Pauchet Y, Muck A, Svatoš A, Heckel DG, Preiss S (2008) Mapping the larval midgut lumen proteome of Helicoverpa armigera, a generalist herbivorous insect. J Proteome Res 7(4):1629–1639. https://doi.org/10.1021/pr7006208
Phugare SS, Kalyani DC, Gaikwad YB, Jadhav JP (2013) Microbial degradation of imidacloprid and toxicological analysis of its biodegradation metabolites in silkworm (Bombyx mori). Chem Eng J 230:27–35
Priya Bhaskaran KP, Bindu PU, Akhilesh VP, Rukhsana K, Jisha Krishnan EK, Sebastian CD (2015) Profiling of catalase and hydrogen peroxide activity in tryptophan administered final instar larvae of Bombyx mori L. Int J Pure App Biosci 3(3):201–207
Quinlan CL, Orr AL, Perevoshchikova IV, Treberg JR, Ackrell BA, Brand MD (2012) Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. J Biol Chem 287(32):27255–27264. https://doi.org/10.1074/jbc.M112.374629
Reese AT, Cho EH, Klitzman B, Nichols SP, Wisniewski NA, Villa MM, Durand HK, Jiang S, Midani FS, Nimmagadda SN, O'Connell TM, Wright JP, Deshusses MA, David LA (2018) Antibiotic-induced changes in the microbiota disrupt redox dynamics in the gut. ELIFE 7:e35987. https://doi.org/10.7554/eLife.35987
Robinson PK (2015) Enzymes: principles and biotechnological applications. Essays Biochem 59:1–41. https://doi.org/10.1042/bse0590001
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Sharma KK, Kuhad RC (2009) An evidence of laccases in archaea. Ind J Microbiol 49:142–150. https://doi.org/10.1007/s12088-009-0039-4
Sharma KK, Singh D, Rawat S (2018) Molecular dynamics simulation studies suggests unconventional roles of non-secretary laccases from enteropathogenic gut bacteria and Cryptococcus neoformans serotype D. Comput Biol Chem 73:41–48. https://doi.org/10.1016/j.compbiolchem.2018.01.010
Shinde AA, Shaikh FK, Gadge PP, Padul MV, Govindwar SP, Kachole MS (2019) Conserved nature of Helicoverpa armigera gut bacterial flora on different host plants and in vitro interactions with PI proteins advocates role in host digestive physiology. J Saudi Soc Agric Sci 18(2):141–149
Singh D, Rawat S, Waseem M, Gupta S, Lynn A, Nitin M, Ramchiary N, Sharma KK (2016) Molecular modeling and simulation studies of recombinant laccase from Yersinia enterocolitica suggests significant role in the biotransformation of non-steroidal anti-inflammatory drugs. Biochem Biophys Res Commun 469(2):306–312. https://doi.org/10.1016/j.bbrc.2015.11.096
Singh D, Sharma KK, Dhar MS, Virdi JS (2014) Molecular modeling and docking of novel laccase from multiple serotype of Yersinia enterocolitica suggests differential and multiple substrate binding. Biochem Biophys Res Commun 449:157–162. https://doi.org/10.1016/j.bbrc.2014.05.003
Sinitcyn P, Rudolph JD, Cox J (2018) Computational methods for understanding mass spectrometry–based shotgun proteomics data. Annu Rev Biomed Data Sci 1:207–234. https://doi.org/10.1146/annurev-biodatasci-080917-013516
Solís-Oba M, Ugalde-Saldívar VM, González I, Viniegra-González V (2005) An electrochemical–spectrophotometrical study of the oxidized forms of the mediator 2, 2′-azino-bis-(3-ethylbenzothiazoline-6-sulfonic acid) produced by immobilized laccase. J Electroanal Chem 579:59–66. https://doi.org/10.1016/j.jelechem.2005.01.025
Sørensen HP, Mortensen KK (2005) Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb Cell Fact 4(1):1. https://doi.org/10.1186/1475-2859-4-1
Spanier B, Starke M, Higel F, Scherer S, Fuchs TM (2010) Yersinia enterocolitica infection and tcaA-dependent killing of Caenorhabditis elegans. Appl Environ Microbiol 76(18):6277–6285. https://doi.org/10.1128/AEM.01274-10
Sun Z, Xu C, Chen S, Shi Q, Wang H, Wang R et al (2019) Exposure to herbicides prime P450-mediated detoxification of Helicoverpa armigera against insecticide and fungal toxin. Insects 10(1):28
Suwanchaichinda C, Sun D, Brattsten LB (2014) Identification and analysis of NADPH-cytochrome p450 reductase in aedes sollicitans (Diptera: Culicidae). J Med Entomol 51(5):958–963. https://doi.org/10.1603/ME13072
Tsai CJY, Loh JMS, Proft T (2016) Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7(3):214–229
Tsigaridas A, Papanikolaou IS, Vaiopoulou A, Anagnostopoulos AK, Viazis N, Karamanolis G et al (2017) Proteomics and irritable bowel syndrome. Expert Rev Proteomics 14(5):461–468. https://doi.org/10.1080/14789450.2017.1317600
Vera A, González-Montalbán N, Arís A, Villaverde A (2007) The conformational quality of insoluble recombinant proteins is enhanced at low growth temperatures. Biotechnol Bioeng 96(6):1101–1106. https://doi.org/10.1002/bit.21218
Virdi JS, Kumar P, Malik S, Bhagat N, Gulati P (2012) Insights into the genetic relationships between environmental and clinical strains of Yersinia enterocolitica Biovar 1A. In: Satyanarayana T, et al. (eds) Microorganisms in environmental management: microbes and environment. Springer Science, Berlin, pp 61–80. https://doi.org/10.1007/978-94-007-2229-3_3
Wang J, Weng Q, Hu Q (2019) Effects of destruxin A on silkworm’s immunophilins. Toxins 11(6):349. https://doi.org/10.3390/toxins11060349
Wang T, Geng SL, Guan YM, Xu WH (2018) Deacetylation of metabolic enzymes by Sirt2 modulates pyruvate homeostasis to extend insect lifespan. Aging (Albany NY) 10(5):1053. https://doi.org/10.18632/aging.101447
Wei D, Zhang MY, Zhang YX, Zhang SY, Smagghe G, Wang JJ (2019) Reduced glutamine synthetase activity alters the fecundity of female Bactrocera dorsalis (Hendel). Insects 10(7):186. https://doi.org/10.3390/insects10070186
Wills ED (1966) Mechanisms of lipid peroxide formation in animal tissues. Biochem J 99:667–676. https://doi.org/10.1042/bj0990667
Zhang G, Zhang W (2018) Protein-protein interaction network analysis of insecticide resistance molecular mechanism in Drosophila melanogaster. Arch Insect Biochem Physiol. https://doi.org/10.1002/arch.21523
Zhu X, Williamson PR (2004) Role of laccase in the biology and virulence of Cryptococcus neoformans. FEMS Yeast Res. https://doi.org/10.1016/j.femsyr.2004.04.004
Acknowledgements
The authors thank Prof. Deepak Pental, CGMCP, University of Delhi South Campus, New Delhi, India for the insect trial facility.
Supplementary information
Supplementary Figure 1—Optimization of recombinant laccase production from Yersinia enterocolitica strain 8081.
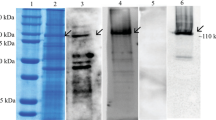
Supplementary Figure 2—I Coomassie-stained SDS-PAGE gel of induced E. coli BL21 cells harboring expression plasmid vector pET28a with laccase gene from Y. enterocolitica strain 8081. Induction was done at 200 rpm with 1 mM IPTG. Lanes: a Control (without induction) b Induction for 2 h at 25 °C; c Induction for 3 h at 25 °C; d Induction for 4 h at 25 °C; e Induction for 2 h at 30 °C; f Induction for 3 h at 30 °C; g Induction for 4 h at 30 °C; h Induction for 2 h at 37 °C; i Induction for 3 h at 37 °C; j Induction for 4 h at 37 °C. II Expression at 100 rpm for 4 h and then kept at static for 16 h at 30 °C a–d: from a Induction at 30 °C; b Induction at 25 °C; c Induction at 16 °C and d uninduced III Expression at 100 rpm for 4 h and then kept at static for 16 h at 4 °C. a Induction at 25 °C; b. Induction at 16 °C.
Supplementary Figure 3—A Coomassie-stained SDS-PAGE gel of solubilized inclusion bodies: I Supernatant II Pellet B & C. Purification of laccase protein using B. Ni–NTA affinity chromatography: I Wash (I) II Wash (II) and III Eluted laccase C Zymogram of purified laccase using ABTS as substrate
Supplementary Figure 4—Cyclic voltammograms obtained for Yersinia enterocolitica strain 8081 (A, B, C) and Bacillus pumilis strain DSKK1 (D, E, F) recombinant laccases with 1mM ABTS (in red) and without ABTS (in blue) A, D at pH 5 using citrate-phosphate buffer; B, E at pH 7 using Tris buffer, and C, F at pH 10 using carbonate-bicarbonate buffer.
Supplementary Table 1—Composition of artificial diet
Supplementary Table 2—Gene ontology (GO) analysis of identified proteins (≥ 1 PSMs in black and ≥ 5 PSMs in blue) from H. armigera larvae with and without recombinant laccase feeding
Supplementary Table 3—Distribution of differentially regulated proteins (with PSMs ≥ 5) from normal diet-fed and recombinant laccase-fed H. armigera larvae gut into various enzyme classes
Funding
This work is supported by grants from the UGC-New Delhi [Grant No. 39-204/1010(SR)]. The FIST-DST grant to the Department of Microbiology is sincerely acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Research involving animal and/or human rights
This article does not contain any studies with animals performed by any of the authors. Helicoverpa armigera has not been notified under any act or laws and rules thereof of the Government of India as an endangered or threatened species restricting or regulating its collection and observation. Therefore, no permits were required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ahlawat, S., Singh, D., Yadav, A. et al. Proteomic analysis reveals the damaging role of low redox laccase from Yersinia enterocolitica strain 8081 in the midgut of Helicoverpa armigera. Biotechnol Lett 42, 2189–2210 (2020). https://doi.org/10.1007/s10529-020-02925-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-020-02925-x