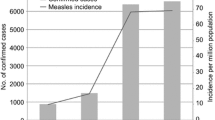
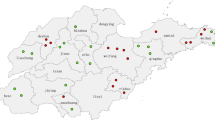
Abstract
We previously reported genotype D11 strains of measles virus that were first isolated from a measles outbreak associated with imported cases in Yunnan province of China by Zhang et al. (Emerg Infect Dis 16(6):943–7, 2010). Genotype D11 has been identified as the 24th genotype of the WHO reference strains. In this study, we sequenced the whole genome of a D11 strain. Phylogenetic analysis using the complete genome sequences of D11 and other reference strains showed that the D11 strain formed a distinct branch that was distant from the other genotypes and was most closely related to the reference strain D7. The M-F non-coding region (NCR) and the N450 coding region sequence (CDS) were found to be the most variable regions. This report provides basic genetic data on genotype D11 for further study of measles evolution and the support for measles elimination.

Similar content being viewed by others
References
Moss WJ, Griffin DE (2012) Measles. Lancet 379(9811):153–164. https://doi.org/10.1016/s0140-6736(10)62352-5
Rima B, Collins P, Easton A, Fouchier R, Kurath G, Lamb RA et al (2017) ICTV virus taxonomy profile: pneumoviridae. J Gen Virol 98(12):2912–2913. https://doi.org/10.1099/jgv.0.000959
Moss WJ (2017) Measles. Lancet 390(10111):2490–2502. https://doi.org/10.1016/s0140-6736(17)31463-0
Rota PA, Brown K, Mankertz A, Santibanez S, Shulga S, Muller CP et al (2011) Global distribution of measles genotypes and measles molecular epidemiology. J Infect Dis 204(Suppl 1):S514–S523. https://doi.org/10.1093/infdis/jir118
Zhang Y, Ding Z, Wang H, Li L, Pang Y, Brown KE et al (2010) New measles virus genotype associated with outbreak, China. Emerg Infect Dis 16(6):943–947. https://doi.org/10.3201/eid1606.100089
Rota PA, Bankamp B (2015) Whole-genome sequencing during measles outbreaks. J Infect Dis 212(10):1529–1530. https://doi.org/10.1093/infdis/jiv272
López-Galíndez C, Penedos AR, Myers R, Hadef B, Aladin F, Brown KE (2015) Assessment of the utility of whole genome sequencing of measles virus in the characterisation of outbreaks. PLoS One 10(11):e0143081. https://doi.org/10.1371/journal.pone.0143081
Gardy JL, Naus M, Amlani A, Chung W, Kim H, Tan M et al (2015) Whole-genome sequencing of measles virus genotypes H1 and D8 during outbreaks of infection following the 2010 olympic winter games reveals viral transmission routes. J Infect Dis 212(10):1574–1578. https://doi.org/10.1093/infdis/jiv271
World Health Organization (2018) The role of extended and whole genome sequencing for tracking transmission of measles and rubella viruses report from the global measles and rubella laboratory network meeting, 2017. Wkly Epidemiol Rec 2018(93):45–60
Bankamp B, Hodge G, McChesney MB, Bellini WJ, Rota PA (2008) Genetic changes that affect the virulence of measles virus in a rhesus macaque model. Virology 373(1):39–50. https://doi.org/10.1016/j.virol.2007.11.025
Parks CL, Lerch RA, Walpita P, Wang HP, Sidhu MS, Udem SA (2001) Comparison of predicted amino acid sequences of measles virus strains in the Edmonston vaccine lineage. J Virol 75(2):910–920. https://doi.org/10.1128/jvi.75.2.910-920.2001
Acknowledgements
The authors acknowledge the staff of the provincial measles laboratory of the Yunnan Center for Disease Control and Prevention for providing the D11 isolate.
Funding
This work was supported by the Key Technologies R&D Program of the National Ministry of Science (2017ZX10104001002, 2018ZX10713002, 2018ZX10201002-006, 2018ZX10711001, 2018ZX10713001-003).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Handling Editor: Bert K. Rima.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, H., Song, J., Xu, W. et al. Whole-genome sequence analysis of the 24th genotype D11 of measles virus. Arch Virol 165, 1895–1898 (2020). https://doi.org/10.1007/s00705-020-04671-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04671-x