Abstract
Although sunflower (Helianthus annuus L.) is categorized as a medium drought-sensitive crop, in a changing climate scenario and/or with the onset of early droughts, the crop may be affected by water stress. This study characterizes and compares the gene expression profiles between aerial part and roots of two sunflower inbred lines with contrasting response to water stress: B59 (sensitive) and B71 (tolerant) under non-stress and water stress. Microarray analysis revealed that water stress induced significant changes in gene expression of both lines. The B59 line had a higher number of genes differentially expressed in water-stressed seedlings compared with B71 line. In both lines, most of the water stress responding genes was upregulated. In B59, the number of genes specifically upregulated in aerial part was higher than that observed in B71. In roots, B71 had more upregulated genes compared with B59. Genes involved in signaling pathway of hormones, components of redox system, and secondary metabolites were enriched in both organs of two lines. The knowledge generated could be helpful to provide tools, that together with the genetic engineering and molecular breeding, it would contribute for the development of water stress-tolerant varieties in crop plants.









Similar content being viewed by others
References
Abid G, Muhovski Y, Mingeot D, Watillon B, Toussaint A, Mergeai G, M’hamdi M, Sassi K, Jebara M (2015) Identification and characterization of drought stress responsive genes in faba bean (Vicia faba L.) by suppression subtractive hybridization. Plant Cell Tissue Organ Cult 121:367–379
Andrade A, Vigliocco A, Alemano S, Alvarez D, Abdala G (2009) Differential accumulation of abscisic acid and its catabolites in drought-sensitive and drought-tolerant sunflower seeds. Seed Sci Res 19:201–211
Andrade A, Vigliocco A, Alemano S, Llanes A, Abdala G (2013) Comparative morpho-biochemical responses of sunflower lines sensitive and tolerant to water stress. Am J Plant Sci 4:156–167
Andrade A, Escalante M, Vigliocco A, Tordable M, Alemano S (2017) Involvement of OPDA and JA in the sunflower seedling responses to moderate water stress. Plant Growth Regul 83:501–511
Cabello JV, Giacomelli JI, Piattoni CV, Iglesias AA, Chan RL (2016) The sunflower transcription factor HaHB11 improves yield, biomass and tolerance to flooding in transgenic Arabidopsis plants. J Biotechnol 222:73–83
Colebrook EH, Thomas SG, Phillips AL, Hedden P (2014) The role of gibberellin signalling in plant responses to abiotic stress. J Exp Biol 217:67–75
Corti Monzón G, Pinedo M, Di Rienzo J, Novo-Uzal N, Pomar F, Lamattina L, de la Canal L (2014) Nitric oxide is required for determining root architecture and lignin composition in sunflower. Supporting evidence from microarray analyses. Nitric Oxide 39:20–28
Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S (2008) A genetic framework for the control of cell division and differentiation in the root meristem. Science 322:1380–1384
Deng XF, Fu FL, Ni N, Li WC (2009) Differential gene expression in response to drought stress in maize seedling. Agric Sci China 8:767–776
Di Rienzo JA, Casanoves F, Balzarini MG, Gonzalez L, Tablada M, Robledo CW (2013) InfoStat versión 2013. Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina. Available at http://www.infostat.com.ar ()
El-Maarouf-Bouteau H, Sajjad Y, Bazin J, Langlade N, Cristescu SM, Balzergue S, Baudouin E, Bailly C (2015) Reactive oxygen species, abscisic acid and ethylene interact to regulate sunflower seed germination. Plant Cell Environ 38:364–374
Fahad S, Bajwa AA, Nazir U, Anjum SA, Farooq A, Zohaib A, Sadia S, Nasim W, Adkins S, Saud S, Ihsan MZ, Alharby H, Wu C, Wang D, Huang J (2017) Crop production under drought and heat stress: plant responses and management options. Front Plant Sci 8:1147
Fang Y, Xiong L (2015) General mechanisms of drought response and their application in drought resistance improvement in plants. Cell Mol Life Sci 72:673–689
Fernández P, Di Rienzo JA, Fernandez L, Hopp HE, Paniego N, Heinz RA (2008) Transcriptomic identification of candidate genes involved in sunflower responses to chilling and salt stresses based on cDNA microarray analysis. BMC Plant Biol 8:11
Fernández P, Soria M, Blesa D, Di Rienzo JA, Moschen S, Peluffo L, Rivarola M, González S, Clavijo B, Principi D (2012) Development, characterization and experimental validation of a cultivated sunflower (Helianthus annuus L.) gene expression oligo microarray. PLoS One 7:1–11
Fracasso A, Trindade LM, Amaducci S (2016) Drought stress tolerance strategies revealed by RNA-Seq in two sorghum genotypes with contrasting WUE. Plant Biol 16:115
Grondin A, Mauleon R, Vadez V, Henry A (2016) Root aquaporins contribute to whole plant water fluxes under drought stress in rice (Oryza sativa L.). Plant Cell Environ 39:347–365
Guo P, Baum M, Grando S, Ceccarelli S, Bai G, Li R, von Korff M, Varshney RK, Graner A, Valkoun K (2009) Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. J Exp Bot 60:3531–3544
Guo L, Yang H, Zhang X, Yang S (2013) Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J Exp Bot 64:1755–1767
He G-H, Xu J-Y, Wang Y-X, Liu J-M, Li P-S, Chen M, Ma Y-Z, Xu Z-S (2016) Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol 16:116
Herrera-Vásquez A, Salinas P, Holuigue L (2015) Salicylic acid and reactive oxygen species interplay in the transcriptional control of defense genes expression. Front Plant Sci 6:171
Jain D, Chattopadhyay D (2010) Analysis of gene expression in response to water deficit of chickpea (Cicer arietinum L.) varieties differing in drought tolerance. BMC Plant Biol 10:24
Joshi R, Wani SH, Singh B, Bohra A, Dar ZA, Lone AA, Pareek A, Singla-Pareek SL (2016) Transcription factors and plants response to drought stress: current understanding and future directions. Front Plant Sci 7:1029
Khan MA, Gemenet DC, Villordon A (2016) Root system architecture and abiotic stress tolerance: current knowledge in root and tuber crops. Front Plant Sci 7:1584
Lenka SK, Katiyar A, Chinnusamy V, Bansal KC (2011) Comparative analysis of drought-responsive transcriptome in Indica rice genotypes with contrasting drought tolerance. Plant Biotechnol J 9:315–327
Liang C, Wang W, Wang J, Ma J, Li C, Zhou F, Zhang S, Yu Y, Zhang L, Li W, Huang X (2017) Identification of differentially expressed genes in sunflower (Helianthus annuus L.) leaves and roots under drought stress by RNA sequencing. Bot Stud 58:42
Ling Z, Zhike Z, Shunquan L, Tingting Z, Xianghui Y (2013) Evaluation of six methods for extraction of total RNA from loquat. Not Bot Horti Agrobo 41:313–316
Lorens GF, Bennett JM, Loggale LB (1987) Differences in drought resistance between two corn hybrids. I. Water relations and root length density. Agron J 79:802–807
Ma J, Li M-Y, Wang F, Tang J, Xiong A-S (2015) Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genomics 16(1):33
Ma S, Bohnert HJ (2007) Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol 8:R49
Mantione KJ, Kream RM, Kuzelova H, Ptacek R, Raboch J, Samuel JM, Stefano GB (2014) Comparing bioinformatic gene expression profiling methods: microarray and RNA-Seq. Med Sci Monit Basic Res 20:138–141
Mead R, Gilmour S, Mead A (2012) Statistical principles for the design of experiments: applications to real experiments. University Press, Cambridge
Meng S, Zhanga C, Sua L, Lia Y, Zhao Z (2016) Nitrogen uptake and metabolism of Populus simonii in response to PEG-induced drought stress. Environ Exp Bot 123:78–87
Miura K, Tada Y (2014) Regulation of water, salinity, and cold stress responses by salicylic acid. Front Plant Sci 5:4
Mohanty B, Kitazumi A, Maurice Cheunga CY, Lakshmananc M, de los Reyes BG, Jang Y-C, Lee D-Y (2016) Identification of candidate network hubs involved in metabolic adjustments of rice under drought stress by integrating transcriptome data and genome-scale metabolic network. Plant Sci 242:224–239
Moschen S, Bengoa Luoni S, Paniego NB, Hopp HE, Dosio GAA, Fernández P, Heinz RA (2014) Identification of candidate genes associated with leaf senescence in cultivated sunflower (Helianthus annuus L.). PLoS One 9:e104379
Moschen S, Bengoa Luoni LS, Di Rienzo J, Caro M, Tohge T, Watanabe M, Hollmann J, González S, Rivarola M, García-García F, Dopazo J, Hopp HE, Hoefgen R, Fernie A, Paniego N, Fernández P, Heinz R (2016a) Integrating transcriptomic and metabolomic analysis to understand natural leaf senescence in sunflower. Plant Biotechnol J 14:719–734
Moschen S, Higgins J, Di Rienzo JA, Heinz RA, Paniego N, Fernández P (2016b) Network and biosignature analysis for the integration of transcriptomic and metabolomic data to characterize leaf senescence process in sunflower. BMC Bioinformatics 17:174
Moschen S, Di Rienzo JA, Higgins J, Tohge T, Watanabe M, González S, Rivarola M, García-García F, Dopazo J, Hopp HE, Hoefgen R, Fernie AR, Paniego N, Fernández P, Heinz RA (2017) Integration of transcriptomic and metabolic data reveals hub transcription factors involved in drought stress response in sunflower (Helianthus annuus L.). Plant Mol Biol 94:549–564
Moumeni A, Satoh K, Kondoh H, Asano T, Hosaka A, Venuprasad R, Serraj R, Kumar A, Leung H, Kikuchi S (2011) Comparative analysis of root transcriptome profiles of two pairs of drought-tolerant and susceptible rice near-isogenic lines under different drought stress. BMC Plant Biol 11:174
Müller M, Munné-Bosch S (2015) Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol 169:32–41
Muñoz-Espinoza VA, López-Climent MF, Casaretto JA, Gómez-Cadenas A (2015) Water stress responses of tomato mutants impaired in hormone biosynthesis reveal abscisic acid, jasmonic acid and salicylic acid interactions. Front Plant Sci 6:997
Mwale SS, Hamusimbi C, Mwansa K (2003) Germination, emergence and growth of sunflower (Helianthus annuus L.) in response to osmotic seed priming. Seed Sci Technol 31:199–206
Osakabe Y, Osakabe K, Shinozaki K, Tran L-SP (2014) Response of plants to water stress. Front Plant Sci 5:86
Pellegrineschi A, Reynolds M, Pacheco M, Brito RM, Almeraya R, Yamaguchi-Shinozaki K, Hoisington D (2004) Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome 47:493–450
Pierik R, Sasidharan R, Voesenek LACJ (2007) Growth control by ethylene: adjusting phenotypes to the environment. J Plant Growth Regul 26:188–200
Pinheiro J, Bates D, DebRoy S, Sarkar D (2012) R Core Team, nlme: linear and nonlinear mixed effects models, R package version 3, 1–104
R Core Team (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at http://www.R-project.org/
Rajala A, Peltonen-Sainio P (2001) Plant growth regulator effects on spring cereal root and shoot growth. Agron J 93:936–943
Rauf S (2008) Breeding sunflower (Helianthus annuus L.) for drought tolerance. Commun Biom Crop Sci 3:29–44
Rengel D, Arribat S, Maury P, Martin-Magniette ML, Hourlier T, Laporte M, Varès D, Carrère S, Grieu P, Balzergue S, Gouzy J, Vincourt P, Langlade NB (2012) A gene phenotype network based on genetic variability for drought responses reveals key physiological processes in controlled and natural environments. PLoS One 7:e45249
Roche J, Hewezi T, Bouniols A, Gentzbittel L (2007) Transcriptional profiles of primary metabolism and signal transduction-related genes in response to water stress in field-grown sunflower genotypes using a thematic cDNA microarray. Planta 226:601–617
Rowe JH, Topping JF, Liu J, Lindsey K (2016) Abscisic acid regulates root growth under osmotic stress conditions via an interacting hormonal network with cytokinin, ethylene and auxin. New Phytol 211:225–239
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Ruzicka K, Ljung K, Vanneste S, Podhorska R, Beeckman T, Friml J, Benková E (2007) Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19:2197–2212
Sah SK, Reddy KR, Li J (2016) Abscisic acid and abiotic stress tolerance in crop plants. Front Plant Sci 7:571
Sahi C, Singh A, Kumar K, Blumwald E, Grover A (2006) Salt stress response in rice: genetics, molecular biology, and comparative genomics. Funct Integr Genomics 6:263–284
Sarazin V, Duclercq J, Guillot X, Sangwan B, Sangwan RS (2017) Water-stressed sunflower transcriptome analysis revealed important molecular markers involved in drought stress response and tolerance. Environ Exp Bot 142:45–63
Shin JH, Vaughn JN, Abdel-Haleem H, Chavarro C, Abernathy B, Kim KD, Jackson SA, Li Z (2015) Transcriptomic changes due to water deficit define a general soybean response and accession-specific pathways for drought avoidance. BMC Plant Biol 15:26
Seo M, Aoki H, Koiwai H, Kamiya Y, Nambara E, Koshiba T (2004) Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol 45:1694–1703
Smyth GK (2005) Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W (eds) Bioinformatics and computational biology solutions using R and bioconductor. Springer, New York, pp 397–420
Stolf-Moreira R, Lemos E, Carareto-Alves L, Marcondes J, Pereira S, Rolla A, Pereira R, Neumaier N, Binneck E, Abdelnoor R, de Oliveira M, Marcelino F, Farias J, Nepomuceno A (2011) Transcriptional profiles of roots of different soybean genotypes subjected to drought stress. Plant Mol Biol Report 29:19–34
Tapia G, Morales-Quintana L, Parra C, Berbel A, Alcorta M (2013) Study of nsLTPs in Lotus japonicus genome reveal a specific epidermal cell member (LjLTP10) regulated by drought stress in aerial organs with a putative role in cutin formation. Plant Mol Biol 82:485–501
Thimm O, Essigmann B, Kloska S, Altmann T, Buckhout TJ (2001) Response of Arabidopsis to iron deficiency stress as revealed by microarray analysis. Plant Physiol 127:1030-1043
Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37:914–939
Ullah A, Sun H, Yang X, Zhang X (2017) Drought coping strategies in cotton: increased crop per drop. Plant Biotechnol J 15:271–284
Verma V, Ravindran P, Kumar PP (2016) Plant hormone-mediated regulation of stress responses. BMC Plant Biol 16:86
Wang X, Cai X, Xu C, Wang Q, Dai S (2016) Drought-responsive mechanisms in plant leaves revealed by proteomics. Int J Mol Sci 17:1706
Wang H, Zhao S, Gao Y, Yang J (2017) Characterization of Dof transcription factors and their responses to osmotic stress in poplar (Populus trichocarpa). PLoS One 12:e0170210
Wilkinson S, Davies WJ (2010) Drought, ozone, ABA and ethylene: new insights from cell to plant to community. Plant Cell Environ 33:510–525
Ye T, Shi S, Wang Y, Chan Z (2015) Contrasting changes caused by drought and submergence stresses in bermudagrass (Cynodon dactylon). Front Plant Sci 6:951
Zhao P, Liu P, Yuan G, Jia J, Li X, Qi D, Chen S, Ma T, Liu G, Cheng L (2016) New insights on drought stress response by global investigation of gene expression changes in sheepgrass (Leymus chinensis). Front Plant Sci 7:954
Zhao S, Fung-Leung W-P, Bittner A, Ngo K, Liu X (2014) Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One 9(1):e78644
Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136:2621–2632
Acknowledgements
Dr. Julia Sabio y Garcia is gratefully acknowledged for critical reading of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
• Microarray revealed that water stress induced significant changes in gene expression of sunflower inbred lines.
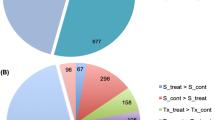
• In B59, the genes upregulated in aerial part were higher than in B71. In roots, B71 had more upregulated genes compared with B59.
• Genes involved in signaling pathway of hormones, components of redox system, and secondary metabolites were enriched in both organs of two lines.
• The identification of unique genes may be used as candidate for selection of water stress tolerance traits
Electronic Supplementary Material
Supplementary Table 1
Description of differentially expressed genes (DEGs) in Aerial Part (AP) of B59 line in water stress vs. control contrast. (XLSX 404 kb)
Supplementary Table 2
Description of differentially expressed genes (DEGs) in Aerial Part (AP) of B71 line in water stress vs. control contrast. (XLSX 281 kb)
Supplementary Table 3
Description of differentially expressed genes (DEGs) in Roots (R) of B59 line in water stress vs. control contrast. (XLSX 216 kb)
Supplementary Table 4
Description of differentially expressed genes (DEGs) in Roots (R) of B71 line in water stress vs. control contrast. (XLSX 262 kb)
Supplementary Table 5
Primer sequences for gene expression analysis derived from RT -qPCR analysis. (30 kb)
Supplementary Table 6
Description of unique differentially expressed genes (DEGs) up-regulated in Aerial Part (AP) of B59 line in water stress vs. control contrast. (XLSX 56 kb)
Supplementary Table 7
Description of unique differentially expressed genes (DEGs) down-regulated in Aerial Part (AP) of B59 line in water stress vs. control contrast. (XLSX 62 kb)
Supplementary Table 8
Description of unique differentially expressed genes (DEGs) up-regulated in Aerial Part (AP) of B71 line in water stress vs. control contrast. (XLSX 36 kb)
Supplementary Table 9
Description of unique differentially expressed genes (DEGs) down-regulated in Aerial Part (AP) of B71 line in water stress vs. control contrast. (XLSX 31 kb)
Supplementary Table 10
Description of unique differentially expressed genes (DEGs) up-regulated in Roots (R) of B59 line in water stress vs. control contrast. (XLSX 22 kb)
Supplementary Table 11
Description of unique differentially expressed genes (DEGs) down-regulated in Roots (R) of B59 line in water stress vs. control contrast. (XLSX 26 kb)
Supplementary Table 12
Description of unique differentially expressed genes (DEGs) up-regulated in Roots (R) of B71 line in water stress vs. control contrast. (XLSX 35 kb)
Supplementary Table 13
Description of unique differentially expressed genes (DEGs) down-regulated in Roots (R) of B71 line in water stress vs. control contrast. (XLSX 39 kb)
Supplementary Figure 1
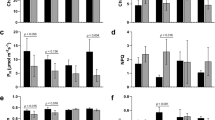
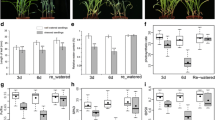
Validation of microarrays of parental lines B71 and B59 under conditions of water stress. Log2 (FC) of the ten selected genes. Elongation factor (Ha.EF1α) and α-TUB were used as reference genes. (JPG 786 kb)
Rights and permissions
About this article
Cite this article
Escalante, M., Vigliocco, A., Moschen, S. et al. Transcriptomic Analysis Reveals a Differential Gene Expression Profile Between Two Sunflower Inbred Lines with Different Ability to Tolerate Water Stress. Plant Mol Biol Rep 38, 222–237 (2020). https://doi.org/10.1007/s11105-020-01192-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-020-01192-4