Abstract
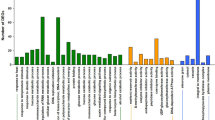
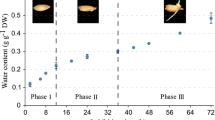
Seed aging is a complex and irreversible process during seed development and storage. The quality of parental seeds affects yield and quality of hybrid rice seeds directly. However, little is known about the mechanism of the accelerated aging seeds in hybrid rice photoperiod-thermo-sensitive genic male sterile (PTGMS) lines. RNA-Seq technique was performed by using seed embryos of two PTGMS lines, Y58S (YS) and Zhun S (ZS), under the restrictive conditions (95% relative humidity and 45 °C) for 0 day, 5 days, 7 days, and 10 days. In total, 1198, 487, 1308, 396, and 669 differentially expressed genes (DEGs) were identified between adjacent time periods of accelerated aging treatment in the ZS and YS. Gene Ontology (GO) analysis revealed the DEGs enriched in the starch catabolic process and sucrose catabolic process. It also showed that many genes were enriched in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways such as starch and sucrose metabolism, plant-pathogen interaction, and plant hormone signal transduction. In addition, 34 candidate genes were selected to the following phylogenetic analysis and different gene expression profiles. Finally, several key candidate genes of seed aging were identified, containing a rice sucrose synthase Os07g0616800 and an unknown gene Os12g0161500, by quantitative real-time PCR. Together, these results established a good foundation for studying the molecular mechanism of rice seed storability and also provide new insights into the PTMGS line seed storage.









Similar content being viewed by others
Abbreviations
- ABA:
-
abscisic acid
- AKR:
-
analdo-ketoreductase
- DEGs:
-
differentially expressed genes
- FC:
-
fold change
- FPKM:
-
fragments per kilobase of transcript per million fragments
- GC:
-
guanine + cytosine
- GO:
-
Gene Ontology
- Hsfs:
-
heat shock transcription factors
- IsoAsp:
-
isoaspartyl
- JA:
-
jasmonic acid
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- PIMT:
-
protein-l-isoaspartyl methyltransferase
- PTGMS:
-
photoperiod-thermo-sensitive genic male sterile
- qRT-PCR:
-
quantitative real-time polymerase chain reaction
- QTL:
-
quantitative trait loci
- ROS:
-
reactive oxygen species
- TFs:
-
transcription factors
- UDP:
-
uridine 5′-diphosphate
- YS:
-
Y58S
- ZS:
-
Zhun S
References
Almoguera C, Prieto-Dapena P, Díaz-Martín J, Espinosa JM, Carranco R, Jordano J (2009) The HaDREB2 transcription factor enhances basal thermotolerance and longevity of seeds through functional interaction with HaHSFA9. BMC Plant Biol 9:75
Anonymous (1983) Seed vigour testing handbook, p 88
Borisjuk L, Rolletschek H (2009) The oxygen status of the developing seed. New Phytol 182:17–30
Broun P (2004) Transcription factors as tools for metabolic engineering in plants. Curr Opin Plant Biol 7:202–209
Carranco R, Espinosa JM, Prieto-Dapena P, Almoguera C, Jordano J (2010) Repression by an auxin/indole acetic acid protein connects auxin signaling with heat shock factor-mediated seed longevity. Proc Natl Acad Sci 107:21908–21913
Chang Z et al (2016) Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc Natl Acad Sci 113:14145–14150
Chen B, Yin G, Whelan J, Zhang Z, Xin X, He J, Chen X, Zhang J, Zhou Y, Lu X (2019) Composition of mitochondrial complex I during the critical node of seed aging in Oryza sativa. J Plant Physiol 236:7–14
Cheng H, Liu H, Deng Y, Xiao J, Li X, Wang S (2015) The WRKY45-2 WRKY13 WRKY42 transcriptional regulatory cascade is required for rice resistance to fungal pathogen. Plant Physiol 167:1087–1099
Day S, Kaya MD (2008) Relationship between seed size and NaCl on germination, seed vigor and early seedling growth of sunflower (Helianthus annuus L.). Afr J Agric Res 3:787–791
Deng Q, Yuan L (1998) Fertility stability of P(T) GMS lines in rice and its identification techniques. Chinese Journal of Riceence 12:200–206
Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38:W64–W70
Egli DB, TeKrony DM, Heitholt JJ, Rupe J (2005) Air temperature during seed filling and soybean seed germination and vigor. Crop Sci 45:1329
Fu Y, Ahmed Z, Diederichsen A (2015) Towards a better monitoring of seed ageing under ex situ seed conservation. Conservation Physiology 3:v26
Fu C et al (2017) Transcriptomic analysis reveals new insights into high-temperature-dependent glume-unclosing in an elite rice male sterile line. Front Plant Sci 8
Gantet P, Memelink J (2002) Transcription factors: tools to engineer the production of pharmacologically active plant metabolites. Trends Pharmacol Sci 23:563–569
Gao J et al (2016) Comparative proteomic analysis of seed embryo proteins associated with seed storability in rice (Oryza sativa L.) during natural aging. Plant Physiol Biochem 103:31–44
Gayen D, Ali N, Sarkar SN, Datta SK, Datta K (2015) Down-regulation of lipoxygenase gene reduces degradation of carotenoids of golden rice during storage. Planta 242:353–363
Gelin JR, Elias EM, Kianian SF (2006) Evaluation of two durum wheat (Triticum turgidum L. var. durum) crosses for pre-harvest sprouting resistance. Field Crop Res 97:188–196
Guo H et al (2017) Transcriptome analysis of neo-tetraploid rice reveals specific differential gene expressions associated with fertility and heterosis. Sci Rep 7:40139
Haigler CH, Ivanova-Datcheva M, Hogan PS, Salnikov VV, Hwang S, Martin K, Delmer DP (2001) Carbon partitioning to cellulose synthesis. Plant Mol Biol 47:29–51
Hamberger B, Ellis M, Friedmann M, de Azevedo SC, Barbazuk B, Douglas CJ (2007) Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: the Populus lignin toolbox and conservation and diversification of angiosperm gene families. Can J Bot 85:1182–1201
Hang NT et al (2015) Mapping QTLs related to rice seed storability under natural and artificial aging storage conditions. Euphytica 203:673–681
Hesse H, Willmitzer L (1996) Expression analysis of a sucrose synthase gene from sugar beet (Beta vulgaris L.). Plant Mol Biol 30:863–872
Hua-Min SI, Liu WZ, Ya-Ping FU, Sun ZX, Guo-Cheng HU (2011) Current situation and suggestions for development of two-line hybrid rice in China. Chin J Rice Sci
Huang C, Lo P, Huang L, Wu S, Yeh C, Lu C (2015) A single-repeat MYB transcription repressor, MYBH, participates in regulation of leaf senescence in Arabidopsis. Plant Mol Biol 88:269–286
Jiang L et al (2011a) Synthetic spike-in standards for RNA-seq experiments. Genome Res 21:1543–1551
Jiang W, Lee J, Jin YM, Qiao Y, Piao R, Jang SM, Woo MO, Kwon SW, Liu X, Pan HY, du X, Koh HJ (2011b) Identification of QTLs for seed germination capability after various storage periods using two RIL populations in rice. Molecules and Cells 31:385–392
Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M (2013) Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42:D199–D205
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360
King SP, Lunn JE, Furbank RT (1997) Carbohydrate content and enzyme metabolism in developing canola siliques. Plant Physiol 114:153–160
Kitin P, Voelker SL, Meinzer FC, Beeckman H, Strauss SH, Lachenbruch B (2010) Tyloses and phenolic deposits in xylem vessels impede water transport in low-lignin transgenic poplars: a study by cryo-fluorescence microscopy. Plant Physiol 154:887–898
Kotak S, Vierling E, Baumlein H, Koskull-Doring PV (2007) A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. THE PLANT CELL ONLINE 19:182–195
LI L et al (2012) Identification of quantitative trait loci for seed storability in rice (Oryza sativa L.). Plant Breed 131:739–743
Li Y, Wang Y, Wang X, Xue H, Pritchard HW (2017) Changes in the mitochondrial protein profile due to ROS eruption during ageing of elm (Ulmus pumila L.) seeds. Plant Physiol Biochem 114:72–87
Lin Q et al (2015) Genetic dissection of seed storability using two different populations with a same parent rice cultivar N22. Breed Sci 65:411–419
Liu C, Mao B, Ou S, Wang W, Liu L, Wu Y, Chu C, Wang X (2014) OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol Biol 84:19–36
Liu SJ, Xu HH, Wang WQ, Li N, Wang WP, Lu Z, Møller IM, Song SQ (2016) Identification of embryo proteins associated with seed germination and seedling establishment in germinating rice seeds. J Plant Physiol 196-197:79–92
Liu S, Liu Y, Jia Y, Wei J, Wang S, Liu X, Zhou Y, Zhu Y, Gu W, Ma H (2017) Gm1-MMP is involved in growth and development of leaf and seed, and enhances tolerance to high temperature and humidity stress in transgenic Arabidopsis. Plant Sci 259:48–61
Luo D, Xu H, Liu Z, Guo J, Li H, Chen L, Fang C, Zhang Q, Bai M, Yao N, Wu H, Wu H, Ji C, Zheng H, Chen Y, Ye S, Li X, Zhao X, Li R, Liu YG (2013) A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat Genet 45:573–577
McDonald MB (1999) Seed deterioration: physiology, repair and assessment. SeedSci. Technol, In, pp 177–237
Min CW et al (2017) In-depth proteomic analysis of Glycine max seeds during controlled deterioration treatment reveals a shift in seed metabolism. J Proteome 169:125–135
Mittal D, Chakrabarti S, Sarkar A, Singh A, Grover A (2009) Heat shock factor gene family in rice: genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol Biochem 47:785–795
Miura K, Lin S, Yano M, Nagamine T (2002) Mapping quantitative trait loci controlling seed longevity in rice (Oryza sativa L.). Theor Appl Genet 104:981–986
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621
Narayana NK et al. (2017) Aldo-ketoreductase 1 (AKR1) improves seed longevity in tobacco and rice by detoxifying reactive cytotoxic compounds generated during ageing. Rice 10
Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619
Rajjou L, Lovigny Y, Groot SPC, Belghazi M, Job C, Job D (2008) Proteome-wide characterization of seed aging in Arabidopsis: a comparison between artificial and natural aging protocols. Plant Physiol 148:620–641
Ratajczak E, Małecka A, Ciereszko I, Staszak AM (2019) Mitochondria are important determinants of the aging of seeds. Int J Mol Sci 20:1568
Ren G, Peng M, Tang WJ, Cai-Guo XU, Xing YZ (2005) QTL associated with seed aging in rice. Acta Agron Sin
Sasaki K, Fukuta Y, Sato T (2005) Mapping of quantitative trait loci controlling seed longevity of rice (Oryza sativa L.) after various periods of seed storage. Plant Breed 124:361–366
Schmelz EA et al (2011) Identity, regulation, and activity of inducible diterpenoid phytoalexins in maize. Proc Natl Acad Sci U S A 108:5455–5460
Schwember AR, Bradford KJ (2010) Quantitative trait loci associated with longevity of lettuce seeds under conventional and controlled deterioration storage conditions. J Exp Bot 61:4423–4436
Shankar R, Bhattacharjee A, Jain M (2016) Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Sci Rep 6
Suzuki A, Suzuki T, Tanabe F, Toki S, Washida H, Wu CY, Takaiwa F (1997) Cloning and expression of five myb-related genes from rice seed. Gene 198:393–398
Takano K (1993) Advances in cereal chemistry and technology in Japan
Toyomasu T (2008) Recent advances regarding diterpene cyclase genes in higher plants and fungi. Biosci Biotechnol Biochem 72:1168–1175
Wang S et al (2015a) The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat Genet 47:949–954
Wang Y et al (2015b) Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat Genet 47:944–948
Wang Y, Li Y, Xue H, Pritchard HW, Wang X (2015c) Reactive oxygen species-provoked mitochondria-dependent cell death during ageing of elm (Ulmus pumila L.) seeds. Plant J 81:438–452
Waterworth WM, Bray CM, West CE (2015) The importance of safeguarding genome integrity in germination and seed longevity. J Exp Bot 66:3549–3558
Wei Y, Xu H, Diao L, Zhu Y, Xie H, Cai Q, Wu F, Wang Z, Zhang J, Xie H (2015) Protein repair l-isoaspartyl methyltransferase 1 (PIMT1) in rice improves seed longevity by preserving embryo vigor and viability. Plant Mol Biol 89:475–492
Yin X, He D, Gupta R, Yang P (2015) Physiological and proteomic analyses on artificially aged Brassica napus seed Frontiers in Plant Science 6
Zeng DL, Guo LB, Xu YB, Yasukumi K, Zhu LH, Qian Q (2006) QTL analysis of seed storability in rice. Plant Breed 125:57–60
Zhang FZF et al (2012) Genome-wide gene expression profiling of introgressed indica rice alleles associated with seedling cold tolerance improvement in a japonica rice background. BMC Genomics
Zhang H et al (2013) Mutation in CSA creates a new photoperiod-sensitive genic male sterile line applicable for hybrid rice seed production. Proc Natl Acad Sci 110:76–81
Zhang Y et al (2016) Transcriptome analysis highlights defense and signaling pathways mediated by rice pi21 gene with partial resistance to Magnaporthe oryzae. Frontiers in Plant Science 7
Zhang X et al (2017) Breeding and study of two new photoperiod- and thermo-sensitive genic male sterile lines of polyploid rice (Oryza sativa L.). Scientific Reports 7
Zhang W, Sun P, He Q, Shu F, Deng H (2018) Transcriptome analysis of near-isogenic line provides novel insights into genes associated with panicle traits regulation in rice. PLoS One 13:e199077
Funding
The project has been funded by the National Key Research and Development Program of China (2018YFD0100900), Hunan Provincial Key Laboratory of Rice and Rapeseed Breeding for Disease Resistance Project (SYKB201705), and Crop Cultivation and Farming Foundation for the Talents of Hunan Agricultural University (YXQN2018-9).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key Message
The team found that DEGs identified from seed embryos were clustered into different processes and metabolisms. Meanwhile, several key candidate genes of seed aging were identified, containing Os07g0616800 and Os12g0161500, by quantitative real-time PCR. Together, these results established a good foundation for studying the molecular mechanism of rice seed storability and also provide new insights into the PTMGS line seed storage.
Electronic Supplementary Material
ESM 1
(XLSX 121 kb)
Rights and permissions
About this article
Cite this article
Liu, Y., He, J., Yan, Y. et al. Comparative Transcriptomic Analysis of Two Rice (Oryza sativa L.) Male Sterile Line Seed Embryos Under Accelerated Aging. Plant Mol Biol Rep 38, 282–293 (2020). https://doi.org/10.1007/s11105-020-01198-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-020-01198-y