Abstract
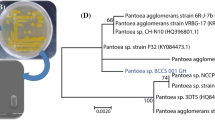
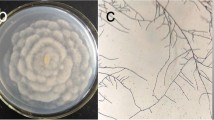
The ascomycete Neodeightonia phoenicum CMIB-151 was isolated from bracts of a palm tree (Syagrus romanzoffiana) in Brazil, and identified by molecular techniques. This is the first report of the isolation of this fungus in South America, and for the first time this fungus has been described as producing an exopolysaccharide (EPS). Different carbon (glucose, fructose, sucrose, maltose, lactose, starch) and nitrogen (yeast extract, peptone, urea, ammonium sulfate) sources were evaluated for the production of EPS. Sucrose and peptone resulted in high production and yield of EPS. High production of EPS occurred at pH 6.0 and 8.0, and acidic conditions (initial pH 5.0) promoted higher mycelial growth. The EPS produced showed a high degree of purity, emulsifying activity and a high water- and oil-holding capacity; important technological properties for industrial applications. FT-IR and 13C-NMR (CP/MAS) analyzes showed typical spectra of carbohydrates containing six-carbon sugar in its structure, and the presence of β-glycosidic configuration with 3-O-substitution. The isolated EPS exhibited remarkable thermal stability.






Similar content being viewed by others
References
Nagi M (2018) Global biopolymers market to witness a cagr of 15.2% during 2018–2024: energias market research Pvt. Ltd. https://www.globenewswire.com/news-release/2018/07/11/1535796/0/en/Global-Biopolymers-Market-to-witness-a-CAGR-of-15-2-during-2018-2024-Energias-Market-Research-Pvt-Ltd.html. Accessed 17 Dec 2019
Cunha MAA, Albornoz SL, Queiroz Santos VA et al (2017) Structure and biological functions of D-glucans and their applications, 1st edn. John Fedor, Amsterdam
Mahapatra M, Banerjee D (2013) Fungal exopolysaccharide: production, composition and applications. Microbiol Insights 6:1–16. https://doi.org/10.4137/MBI.S10957
Kagimura FY, da Cunha MAA, Barbosa AM et al (2015) Biological activities of derivatized D-glucans: a review. Int J Biol Macromol 72:588–598. https://doi.org/10.1016/j.ijbiomac.2014.09.008
Huang WC, Tang IC (2007) Bacterial and yeast cultures-process characteristics, products, and applications. In: Yang S-T (ed) Bioprocessing for value-added products from renewable resources. Elsevier, Dublin, pp 185–223
Xu Y, Cui Y, Yue F et al (2019) Exopolysaccharides produced by lactic acid bacteria and bifidobacteria: structures, physiochemical functions and applications in the food industry. Food Hydrocoll 94:475–499. https://doi.org/10.1016/j.foodhyd.2019.03.032
Almasi T, Jabbari K, Gholipour N et al (2019) Synthesis, characterization, and in vitro and in vivo 68Ga radiolabeling of thiosemicarbazone Schiff base derived from dialdehyde dextran as a promising blood pool imaging agent. Int J Biol Macromol 125:915–921. https://doi.org/10.1016/j.ijbiomac.2018.12.133
Cunha MAA, Santos VAQ, Calegari GC et al (2019) Structure and biological properties of lasiodiplodan: An uncommon fungal exopolysaccharide of the (1→ 6)-β-D-glucan type. In: Cohen E, Merzendorfer H (eds) Extracellular sugar-based biopolymers matrices, 1st edn. Cham, Switzerland, pp 409–432
Wu J-Y (2017) Ultrasound-assisted extraction of polysaccharides from edible and medicinal fungi: Major factors and process kinetics. MOJ Food Process Technol. https://doi.org/10.15406/mojfpt.2017.04.00086
Góes-neto A, Loguercio-leite C (2005) DNA extraction from frozen field-collected and dehydrated herbarium fungal basidiomata performance of SDS and CTAB-based methods. Biotemas 18:19–32. https://doi.org/10.5007/%25x
Dentinger BTM, Margaritescu S, Moncalvo J-M (2009) Rapid and reliable high-throughput methods of DNA extraction for use in barcoding and molecular systematics of mushrooms. Mol Ecol Resour 10:628–633. https://doi.org/10.1111/j.1755-0998.2009.02825.x
Green MR (2012) Molecular cloning this is a free sample of content from molecular cloning: a laboratory manual, 4th edn. John Inglis, New York
Kearse M, Moir R, Wilson A et al (2012) Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinforma Appl 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinforma Appl NOTE 19:1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256. https://doi.org/10.1093/molbev/msn083
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. In: 2010 Gateway Computing Environments Workshop, GCE, pp 1–8
Marques MW, Lima NB, De Morais MA et al (2013) Species of Lasiodiplodia associated with mango in Brazil. Fungal Divers 61:181–193. https://doi.org/10.1007/s13225-013-0231-z
Adamčík S, Cai L, Chakraborty D et al (2015) Fungal biodiversity profiles 1–10. Cryptogam Mycol 36:121–166. https://doi.org/10.7872/crym/v36.iss2.2015.121
Dai DQ, Phookamsak R, Wijayawardene NN et al (2017) Bambusicolous fungi. Fungal Divers. https://doi.org/10.1007/s13225-016-0367-8
Phillips AJL, Alves A, Pennycook SR et al (2008) Resolving the phylogenetic and taxonomic status of dark-spored teleomorph genera in the Botryosphaeriaceae. Persoonia Mol Phylogeny Evol Fungi 21:29–55. https://doi.org/10.3767/003158508X340742
Vu D, Groenewald M, de Vries M et al (2019) Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud Mycol 92:135–154. https://doi.org/10.1016/j.simyco.2018.05.001
Ligoxigakis EK, Markakis EA, Papaioannou IA, Typas MA (2013) First report of palm rot of Phoenix spp. caused by Neodeightonia phoenicum in Greece. Plant Dis. 97:286. https://doi.org/10.1094/PDIS-08-12-0727-PDN
Konta S, Phillips A, Bahkali A et al (2016) Botryosphaeriaceae from palms in Thailand—Barriopsis archontophoenicis sp. nov, from Archontophoenix alexandrae. Mycosphere 7:921–932. https://doi.org/10.5943/mycosphere/si/1b/1
Hyde KD, Konta S, Hongsanan S et al (2016) Botryosphaeriaceae from palms in Thailand II-two new species of Neodeightonia, N. rattanica and N. rattanicola from Calamus (rattan palm). Mycosphere 7:90–961. https://doi.org/10.5943/mycosphere/si/1b/6
Zhang J, Kapli P, Pavlidis P, Stamatakis A (2013) Phylogenetics a general species delimitation method with applications to phylogenetic placements. Bioinformatics 29:2869–2876. https://doi.org/10.1093/bioinformatics/btt499
Puillandre N, Lambert A, Brouillet S, Achaz G (2012) ABGD, automatic barcode gap discovery for primary species delimitation. Mol Ecol 21:1864–1877. https://doi.org/10.1111/j.1365-294X.2011.05239.x
Vogel HJ (1956) A convenient growth medium for Neurospora (Medium N). Microb Genet Bull 13:42–43
Cunha MAA, Turmina JA, Ivanov RC et al (2012) Lasiodiplodan, an exocellular (1→6)-β-D-glucan from Lasiodiplodia theobromae MMPI: production on glucose, fermentation kinetics, rheology and anti-proliferative activity. J Ind Microbiol Biotechnol 39:1179–1188. https://doi.org/10.1007/s10295-012-1112-2
Dubois M, Gilles KA, Hamilton JK et al (1956) Colorimetric method for determination of sugars and related substances. Anal Chem 28:350–356. https://doi.org/10.1021/ac60111a017
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Lobo RE, Gómez MI, Font de Valdez G, Torino MI (2019) Physicochemical and antioxidant properties of a gastroprotective exopolysaccharide produced by Streptococcus thermophilus CRL1190. Food Hydrocoll 96:625–633. https://doi.org/10.1016/j.foodhyd.2019.05.036
Govarthanan M, Kamala-Kannan S, Selvankumar T et al (2019) Effect of blue light on growth and exopolysaccharides production in phototrophic Rhodobacter sp. BT18 isolated from brackish water. Int J Biol Macromol 131:74–80. https://doi.org/10.1016/j.ijbiomac.2019.03.049
Phillips A (2009) Taxonomy, phylogeny, and epitypification of Melanops tulasnei, the type species of Melanops. Fungal Divers 38:155–166
Phillips AJL, Alves A, Abdollahzadeh J et al (2013) The botryosphaeriaceae: genera and species known from culture. Stud Mycol 76:51–167. https://doi.org/10.3114/sim0021
Hyde KD, Hongsanan S, Jeewon R et al (2016) Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 80:1–270. https://doi.org/10.1007/s13225-016-0373-x
Papinutti L (2010) Effects of nutrients, pH and water potential on exopolysaccharides production by a fungal strain belonging to Ganoderma lucidum complex. Bioresour Technol 101:1941–1946. https://doi.org/10.1016/j.biortech.2009.09.076
Kagimura FY, Cunha MAA, Theis TV et al (2015) Carboxymethylation of (1→6)-β-glucan (lasiodiplodan): Preparation, characterization and antioxidant evaluation. Carbohydr Polym 127:390–399. https://doi.org/10.1016/j.carbpol.2015.03.045
Guo MQ, Hu X, Wang C, Ai L (2017) Polysaccharides: structure and solubility. In: Xu Z (ed) Solubility of polysaccharides, 1st edn. InTech, London, p 13
Calegari GC, Vidiany Aparecida Queiroz S, Barbosa-Dekker AM et al (2019) Sulfonated (1→6)-β-D-Glucan (lasiodiplodan). Food Technol Biotechnol 57:490–502. https://doi.org/10.17113/ftb.57.04.19.6264
Capriotti K, Capriotti JA (2012) Dimethyl sulfoxide: history, chemistry, and clinical utility in dermatology. J Clin Aesthet Dermatol 5:24–26
Kang D, Cai Z, Wei Y, Zhang H (2017) Structure and chain conformation characteristics of high acyl gellan gum polysaccharide in DMSO with sodium nitrate. Polymer (Guildf) 128:147–158. https://doi.org/10.1016/j.polymer.2017.09.035
Freitas F, Alves VD, Carvalheira M et al (2009) Emulsifying behaviour and rheological properties of the extracellular polysaccharide produced by Pseudomonas oleovorans grown on glycerol byproduct. Carbohydr Polym 78:549–556. https://doi.org/10.1016/j.carbpol.2009.05.016
Petravić-Tominac V, Zechner-Krpan V, Berković K et al (2011) Rheological properties, water-holding and oil-binding capacities of particulate β-glucans isolated from spent brewer’s yeast by three different procedures. Food Technol Biotechnol 49:56–64
Xu J, Liu W, Yao W et al (2009) Carboxymethylation of a polysaccharide extracted from Ganoderma lucidum enhances its antioxidant activities in vitro. Carbohydr Polym 78:227–234. https://doi.org/10.1016/j.carbpol.2009.03.028
Pavia DL, Lampman GM, Kriz GS, Vyvyan JR (2016) Introdução à espectroscopia—Tradução da 5a. edição norte-americana. Cengage Learning, São Paulo-SP
Tian Y, Zeng H, Xu Z et al (2012) Ultrasonic-assisted extraction and antioxidant activity of polysaccharides recovered from white button mushroom (Agaricus bisporus). Carbohydr Polym 88:522–529. https://doi.org/10.1016/j.carbpol.2011.12.042
Buddana SK, Venkata Naga Varanasi Y, Reddy Shetty P (2015) Fibrinolytic, anti-inflammatory and anti-microbial properties of α-(1→3)-glucans produced from Streptococcus mutans (MTCC 497). Carbohydr Polym 115:152–159. https://doi.org/10.1016/j.carbpol.2014.08.083
Taubner T, Marounek M, Synytsya A (2017) Preparation and characterization of amidated derivatives of alginic acid. Int J Biol Macromol 103:202–207. https://doi.org/10.1016/j.ijbiomac.2017.05.070
Ulu A, Koytepe S, Ates B (2016) Design of starch functionalized biodegradable P(MAA-co-MMA) as carrier matrix for L-asparaginase immobilization. Carbohydr Polym 153:559–572. https://doi.org/10.1016/j.carbpol.2016.08.019
David Ebenezar IJ, Ramalingam S, Ramachandra Raja C, Jobe Prabakar PC (2014) Vibrational Spectroscopic (IR and Raman) analysis and computational investigation (NMR, UV-Visible, MEP and Kubo gap) on L-valinium picrate. J Nanotechnol Adv Mater 2:11–25. https://doi.org/10.12785/jnam/020102
Wang J, Zhang L (2009) Structure and chain conformation of five water-soluble derivatives of a β-D-glucan isolated from Ganoderma lucidum. Carbohydr Res 344:105–112. https://doi.org/10.1016/j.carres.2008.09.024
Sánchez LWN, Santos VAQ, Teixeira SD et al (2018) O-Acetylated (1→6)-β-D-Glucan (lasiodiplodan): Chemical derivatization, characterization and antioxidant activity. J Pharm Pharmacol 6(6):320–332. https://doi.org/10.17265/2328-2150/2018.04.003
Alquini G, Carbonero ER, Rosado FR et al (2004) Polysaccharides from the fruit bodies of the basidiomycete Laetiporus sulphureus (Bull.: Fr.) Murr. FEMS Microbiol Lett 230:47–52. https://doi.org/10.1016/S0378-1097(03)00853-X
Synytsya A, Novák M (2013) Structural diversity of fungal glucans. Carbohydr Polym 92:792–809. https://doi.org/10.1016/j.carbpol.2012.09.077
Smiderle FR, Baggio CH, Borato DG et al (2014) Anti-inflammatory properties of the medicinal mushroom Cordyceps militaris might be related to its linear (1→3)-β-D-glucan. PLoS ONE 9:e110266. https://doi.org/10.1371/journal.pone.0110266
Kong L, Yu L, Feng T et al (2015) Physicochemical characterization of the polysaccharide from Bletilla striata: Effect of drying method. Carbohydr Polym 125:1–8. https://doi.org/10.1016/j.carbpol.2015.02.042
Feng F, Zhou Q, Yang Y et al (2018) Characterization of highly branched dextran produced by Leuconostoc citreum B-2 from pineapple fermented product. Int J Biol Macromol 113:45–50. https://doi.org/10.1016/j.ijbiomac.2018.02.119
Vijeth S, Heggannavar GB, Kariduraganavar YM (2019) Encapsulating wall materials for micro-/nanocapsules. In: Salaün F (ed) Microencapsulation–processes, technologies and industrial applications, 1st edn. IntechOpen, London, pp 1–19
Kavitake D, Delattre C, Devi PB et al (2019) Physical and functional characterization of succinoglycan exopolysaccharide produced by Rhizobium radiobacter CAS from curd sample. Int J Biol Macromol 134:1013–1021. https://doi.org/10.1016/j.ijbiomac.2019.05.050
Acknowledgements
The authors thank Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brazil) for providing scholarships to MSF (PPGTP-UTFPR) and DKSG (PPGFAP/UFSC), and for partial financing of the research. ERDS (Process no. 311158/2018-8) and AGN are supported by CNPq, Brazil. Part of this research is part of the MIND.Funga Project: https://mindfunga.ufsc.br/.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by MSF, MLKM, DKSG, VAQS and MAAC. The first draft of the manuscript was written by MAAC, AGN, ERDS, AMBD and RFHD and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
da Silva Fonseca, M., Marchioro, M.L.K., Guimarães, D.K.S. et al. Neodeightonia phoenicum CMIB-151: Isolation, Molecular Identification, and Production and Characterization of an Exopolysaccharide. J Polym Environ 28, 1954–1966 (2020). https://doi.org/10.1007/s10924-020-01744-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10924-020-01744-5