Abstract
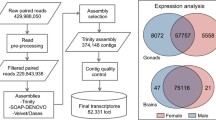
The blood acts as a transfer channel for a variety of factors in the whole body. The ricefield eel (Monopterus albus) is a protogynous hermaphrodite vertebrate. Until now, no research has reported an analysis of the blood transcriptome during the process of sexual development in the ricefield eel. In this study, the transcriptome sequencing of blood samples from male and female ricefield eels was completed with a total of 34.70 Gb clean data. The clean data of each sample all reached 5.23 GB, and the percent of the Q30 basic group was 88.62% and above. A total of 106,369 unigenes were obtained after assembly, including 13,296 unigenes with a length of more than 1 kb. Further functional annotation analysis showed that there are 28,522 unigenes that can be annotated. The annotations of genes with differential expression revealed that there were 563 genes with significant differential expression in the blood of male and female ricefield eels, including 91 upregulated genes and 472 downregulated genes. Among which, 14 genes may be closely related to sex differentiation, the qPCR was used to confirmed the expression pattern of those genes and result shown that 11 genes were downregulated and 3 genes were upregulated, consistent with the results of our RNA-Seq analysis. This blood transcript dataset will open future research avenues on ricefield eel sex development and differentiation.







Similar content being viewed by others
Data availability
Raw sequence data is available on the NCBI Sequence Read Archive under the BioProject Accession number PRJNA388266.
References
Achermann JC, Ito M, Ito M, Hindmarsh PC, Jameson JL (1999) A mutation in the gene encoding steroidogenic factor-1 causes XY sex reversal and adrenal failure in humans. Nat Genet 22:125–126. https://doi.org/10.1038/9629
Cai J, Yang W, Chen D, Zhang Y, He Z, Zhang W, Zhang L (2017) Transcriptomic analysis of the differentiating ovary of the protogynous ricefield eel Monopterus albus. BMC Genomics 18:573. https://doi.org/10.1186/s12864-017-3953-6
Chi W, Gao Y, Hu Q, Guo W, Li D (2017) Genome-wide analysis of brain and gonad transcripts reveals changes of key sex reversal-related genes expression and signaling pathways in three stages of Monopterus albus. PLoS One 12:e0173974. https://doi.org/10.1371/journal.pone.0173974
Deng YY, Li JO, Wu SF, Zhu YP, Chen YW, He FC (2006) Integrated nr database in protein annotation system and its localization. Comput Eng 32:71–74
Doroshov SI, Moberg GP, Van Eenennaam JP (1997) Observations on the reproductive cycle of cultures white sturgeon, Acipenser transmontanus. Environ Biol Fish 48:265–278. https://doi.org/10.1023/a:1007336802423
Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14:755–763
Etebari K, Palfreyman RW, Schlipalius D, Nielsen LK, Glatz RV, Asgari S (2011) Deep sequencing-based transcriptome analysis of Plutella xylostella larvae parasitized by Diadegma semiclausum. BMC Genomics 12:446. https://doi.org/10.1186/1471-2164-12-446
Finn RD et al (2014) Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. https://doi.org/10.1093/nar/gkt1223
Franssen SU, Shrestha RP, Brautigam A, Bornberg-Bauer E, Weber AP (2011) Comprehensive transcriptome analysis of the highly complex Pisum sativum genome using next generation sequencing. BMC Genomics 12:227. https://doi.org/10.1186/1471-2164-12-227
Freedman JE (2011) A platelet transcriptome revolution. Blood 118:3760–3761. https://doi.org/10.1182/blood-2011-05-356600
Grabherr MG et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. https://doi.org/10.1038/nbt.1883
Jiang Y, Han K, Chen S, Hong W, Wang Y, Zhang Z (2015) Molecular cloning, characterization and expression of Lc-Sox11a in large yellow croaker Larimichthys crocea. Gene 574:287–301. https://doi.org/10.1016/j.gene.2015.08.021
Kanai Y, Hiramatsu R, Matoba S, Kidokoro T (2005) From SRY to SOX9: mammalian testis differentiation. J Biochem 138:13–19. https://doi.org/10.1093/jb/mvi098
Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R (1993) WT-1 is required for early kidney development. Cell 74:679–691
Ku HY, Gangaraju VK, Qi H, Liu N, Lin H (2016) Tudor-SN interacts with Piwi antagonistically in regulating spermatogenesis but synergistically in silencing transposons in Drosophila. PLoS Genet 12:e1005813. https://doi.org/10.1371/journal.pgen.1005813
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. https://doi.org/10.1186/gb-2009-10-3-r25
Laterza OF et al (2009) Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem 55:1977–1983. https://doi.org/10.1373/clinchem.2009.131797
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform 12:323. https://doi.org/10.1186/1471-2105-12-323
Liu J, Y GG (1951) The Monopterus albus’s gonal histology change during its sex reversion. Acta Hydrobiol Sinica 2:85–109
Liu H, Lamm MS, Rutherford K, Black MA, Godwin JR, Gemmell NJ (2015) Large-scale transcriptome sequencing reveals novel expression patterns for key sex-related genes in a sex-changing fish. Biol Sex Differ 6:26. https://doi.org/10.1186/s13293-015-0044-8
Matsuba R et al (2015) Replication study in a Japanese population to evaluate the association between 10 SNP loci, identified in European genome-wide association studies, and type 2 diabetes. PLoS One 10:e0126363. https://doi.org/10.1371/journal.pone.0126363
McCabe ER (1996) Sex and the single DAX1: too little is bad, but can we have too much? J Clin Invest 98:881–882. https://doi.org/10.1172/JCI118868
Mitchell PS et al (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 105:10513–10518. https://doi.org/10.1073/pnas.0804549105
Mohr S, Liew CC (2007) The peripheral-blood transcriptome: new insights into disease and risk assessment. Trends Mol Med 13:422–432. https://doi.org/10.1016/j.molmed.2007.08.003
Ohashi T, Sato S, Yoshiki A, Kusakabe M (1997) TSC-36 (follistatin-related polypeptide) gene expression in estrogen receptor positive osteoblastic cell line, CDO7F. Calcif Tissue Int 61:400–403
Schulze SK, Kanwar R, Golzenleuchter M, Therneau TM, Beutler AS (2012) SERE: single-parameter quality control and sample comparison for RNA-Seq. BMC Genomics 13:524. https://doi.org/10.1186/1471-2164-13-524
Taol Z, Xiao HUG, Long-zhenl ZHANG, Fengl ZHAO, Pingl ZHUANG, Bin WANG, Rong FGP (2007) Determination of different sexes and ovarian development stages in cultured Acipenser baerii based on blood biochemical indices. J Fishery Sci China 14:236–243
Trapnell C et al (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. https://doi.org/10.1038/nbt.1621
Uda M et al (2004) Foxl2 disruption causes mouse ovarian failure by pervasive blockage of follicle development. Hum Mol Genet 13:1171–1181. https://doi.org/10.1093/hmg/ddh124
Wei L, Yang C, Tao W, Wang D (2016) Genome-wide identification and transcriptome-based expression profiling of the sox gene family in the Nile Tilapia (Oreochromis niloticus). Int J Mol Sci 17. https://doi.org/10.3390/ijms17030270
Wilhelm Filho D, Eble GJ, Kassner G, Caprario FX, Dafre AL, Ohira M (1992) Comparative hematology in marine fish Comparative biochemistry and physiology. Comp Physiol 102:311–321
Xao YM (1995) Study on the reproductive biology of Monopterus albus (Zuiew) I female development of Monopterusalbus. Acta Sci Nat Univ Norm Hunan 18:45–52
Xao YM, Liu Y (1995) Study on the histology in sex changing from intersex to male of Monopterus ablus. J Fish China 19:297–301
Xie C et al (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39:W316–W322. https://doi.org/10.1093/nar/gkr483
Xu D, Shen KN, Fan Z, Huang W, You F, Lou B, Hsiao CD (2016) The testis and ovary transcriptomes of the rock bream (Oplegnathus fasciatus): a bony fish with a unique neo Y chromosome. Genomics Data 7:210–213. https://doi.org/10.1016/j.gdata.2016.01.007
Yi M, Chen F, Luo M, Cheng Y, Zhao H, Cheng H, Zhou R (2014) Rapid evolution of piRNA pathway in the teleost fish: implication for an adaptation to transposon diversity. Genome Biol Evolut 6:1393–1407. https://doi.org/10.1093/gbe/evu105
Funding
This study was funded by the Central Public-Interest Scientific Institution Basal Research Fund (2015JBFM06).
Author information
Authors and Affiliations
Contributions
WD performed most of the experimental studies and wrote this manuscript.
LC isolated DNA and mRNA and contributed to the PCR analysis.
ZC performed the sequencing and analyses.
XB designed part of the experiments and reviewed the paper.
Corresponding author
Ethics declarations
Ethics approval
Ricefield eel (Monopterus albus) is not an endangered or protected species, and all experiments were approved by the Institutional Animal Care and Use Committee of the Ministry of Freshwater Fisheries Research Center of the Chinese Academy of Fishery Sciences and were undertaken in accordance with the national legislation for fish welfare established by the Ministry of Science and Technology of the People’s Republic of China.
Competing interests
The authors declare that they have no competing interests.
Consent for publish
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Significance statement
A total of 106,369 unigenes were obtained after transcriptome sequencing. The annotations of genes with differential expression revealed that there were 563 genes with significant differential expression in the blood of male and female ricefield eels, including 91 upregulated genes and 472 downregulated genes. Among which, 14 genes may be closely related to sex differentiation, but their specific functions need to be further studied.
Electronic supplementary material
Fig. S1
The gonadal histology of ricefield eel. T01/T02/T03, Spermatophore could be observed in testicle. T04/T05/T06, Oocytes could be observed in ovary. SP:spermatophore; O:ooctytes. (JPG 3013 kb)
Fig. S2
The length distribution of all unigene (JPG 245 kb)
Fig. S3
Phylogenetic tree of nucleotide sequences of potential sex-related gene in ricefield fish along with several other vertebrate species. The tree was estimated by the maximum likelihood method. Each nucleotide sequence at the terminal is presented by the name of species and gene from which the nucleotide sequence originated. The statistical robustness of the tree was estimated by bootstrapping with 1000 replicates. Bar represents the p distance. Words with a yellow background indicate the queried genes related to sex development. a,piwi; b,DNAJB1; c,Uhrf1; d,plcx2; e,FSTL5; f,zmziz1; g,sox11a; h,eomes; i,NASP; j,oocyte zinc finger protein; k,hsp; l,sushi; m,vhsv; n,pfam (PDF 6595 kb)
ESM 1
(DOCX 18 kb)
Rights and permissions
About this article
Cite this article
Ding, W., Cao, L., Cao, Z. et al. Transcriptome analysis of blood for the discovery of sex-related genes in ricefield eel Monopterus albus. Fish Physiol Biochem 46, 1507–1518 (2020). https://doi.org/10.1007/s10695-020-00809-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10695-020-00809-5