Abstract
The purpose of this study was to investigate the prevalence, antibiotic resistance and certain virulence genes of the most predominant bacterial pathogens causing BRD. A total of 225 calves; 55 apparently healthy and 170 diseased; were sampled. Bacteriological examination, antimicrobial susceptibility testing and PCR based detection of some virulence genes were performed. In addition, the serotyping of E. coli was performed using the slide agglutination test. The most predominant bacterial pathogens retrieved from apparently healthy calves were E. coli (16.4%) and S. aureus (10.9%), and in pneumonic calves were E. coli (23.5%), P. vulgaris (12.4%) and S. aureus (11.8%). The most prevalent virulence gene in E. coli was the fimH gene (100%), followed by eaeA gene (24.5%) and hly gene (20.4%). All the examined S. aureus strains harbored spa and coa genes; likewise, all P. multocida strains harbored toxA gene. The majority of the isolated strains displayed remarkable sensitivity to norfloxacin and enrofloxacin; furthermore, the retrieved E. coli strains exhibited multidrug-resistance to gentamicin, erythromycin, streptomycin and trimethoprim-sulphamethoxazole, in addition, the isolated S. aureus and P. aeruginosa strains showed multidrug-resistance to amoxicillin, ampicillin and tetracycline. E. coli serogroups including O18, O143, O1, and O6 were retrieved from pneumonic calves as the first report in Egypt. In conclusion, the synergism between the conventional and genotypic analysis is an effective gadget for the characterization of bacterial pathogens causing BRD. Continuous surveillance of antimicrobial susceptibility is essential to select the drug of choice due to the development of multidrug-resistant strains.
Similar content being viewed by others
Introduction
Bovine respiratory disease (BRD) is one of the major risks to the health of calves and younger cattle, causing significant economic losses (Zeineldin et al. 2017). Viral infections, stresses, nutritional and environmental conditions are leading reasons for the commensal bacteria turned into to be opportunistic pathogens, leading to secondary respiratory infections and increase animal susceptibility to the respiratory disease (Clavijo et al. 2007; Griffin et al. 2010). Many bacteria are involved in BRD, including; Staphylococcus aureus, Pasteurella multocida, Escherichia coli, Citrobacter spp., Pseudomonas aeruginosa, Proteus vulgaris, Enterobacter aerogenes, Enterobacter cloacae, Bacillus spp., and Mannheimia haemolytica (de Oliveira et al. 2016; DebRoy et al. 2008; Timsit et al. 2017; Zeineldin et al. 2017).
Clinical signs of BRD vary from acute to chronic illness and include mostly high fever, depression, loss of appetite, cough, rapid respiratory rates, abnormal lung sounds and nasal discharge (Radostits et al. 2006). The utilization of antibiotics in the treatment of BRD has widely been reported. The emergence of multidrug-resistant pathogens associated with BRD has been recorded that threatens the livestock industry (Klima et al. 2014). The continuous surveillance of antimicrobial susceptibility should be applied as well as the proper use of antibiotics in order to reduce the spread of multidrug-resistant strains not only between livestock but also to humans (Holman et al. 2015; Klima et al. 2014).
The polymerase chain reaction (PCR) is a highly reliable and specific diagnostic tool that facilitates the detection of pathogens and their virulence genes as well as drug resistance genes (Eid et al. 2016; Enany et al. 2018). Therefore, the current work aimed to determine the prevalence of bacterial pathogens in apparently healthy and pneumonic calves and to monitor the antibiotic susceptibility of the isolated strains, and to perform PCR-based detection of certain virulence genes.
Materials and methods
Sampling
A total of 225 calves, in private cattle farms at the Ismailia governorate, Northern Egypt, were examined for respiratory manifestations from November 2018 to January 2019. The examined calves are ranging in age from one day to 6 months old. All calves were clinically examined calves were categorized into 3 groups, according to the clinical signs, the 1st group (n = 154) showed respiratory signs such as fever (> 39.5 °C), rapid respiration, coughing, and nasal discharge, the 2nd group (n = 16) showed only fever, and the 3rd group (apparently healthy; n = 55) showed no signs. Nasal swabs were gathered from 150 calves that exhibited respiratory signs and from 50 apparently healthy calves. Blood samples were collected in ethylenediaminetetraacetic acid (EDTA) from 16 calves that were showed only fever and from 5 apparently healthy calves. Four samples were collected from lung tissues of 4 emergency slaughtered calves. All specimens were collected and transported to the laboratory for further analysis.
Bacteriological examination
The collected samples (nasal swabs and homogenized lung tissue) were inoculated into peptone water followed by incubation at 37 °C for 24 h. A loopful of incubated broth and blood samples were streaked onto Blood agar, Nutrient agar, MacConkey’s agar, Eosin methylene blue agar and Mannitol salt agar (Oxoid, UK), and then incubated at 37 °C for 24-48 h. The characterization of bacterial species was carried out based on the cultural characteristics, Gram’s stain, and biochemical reactions according to the previous report (Quinn et al. 2011).
Serotyping of E. coli
Isolates of E. coli were serotyped based upon somatic and flagellar antigens using the slide agglutination test according to the previous report (Edwards and Ewing 1962). It was performed at Animal Health Research Institute, Dokki, Egypt, using a commercially available antisera kit (pathogenic E. coli immune sera) (Denka SeikenCo., Ltd., Tokyo, Japan). Bacteria showing agglutination within 30 s to antisera were estimated positive for the corresponding serotype.
Pathogenicity test of the isolated P. multocida strains
The P. multocida strains were exposed to the pathogenicity test as the previously described protocol (Varte et al. 2014). Briefly, six mice were I/P inoculated with 100 μl of the whole culture; in addition, three mice were inoculated with 100 μl of sterile H2O (control group). All mice were observed for 2 days. Death within 24-48 h was monitored with postmortem examination. Re-isolation of P. multocida from the heart blood of dead mice was carried out by plating on blood agar. In addition, Giemsa stained smears from heart blood were made and microscopically examined. The handling of mice was carried out as described by the Animal Ethics Committee of Suez Canal University, Egypt.
PCR detection of virulence genes
Genomic DNA of E. coli, S. aureus and P. multocida was extracted according to the manufacturer’s instructions of the QIAamp DNA Mini Kit (Catalogue No. 51304). The PCR reactions were done in a 25-μl volume containing 5 μl of 5× Master Mix (Jena Bioscience, Germany), 20 pmol of each primer and 3 μl of DNA template. Double-distilled H2O was added to complete the reaction volume to 25 μl. Positive controls kindly supplied by Animal Health Research Institute in Dokki, Cairo, Egypt and negative control (no DNA templates) were utilized in all PCRs. The primers sequences (Metabion International AG, Germany) and recycling conditions are listed in Table 1. The PCR products were separated by electrophoresis (1.5% agarose stained with ethidium bromide 0.5 μg/ml), and the amplicon was captured using a gel documentation system (Biospectrum UVP, UK).
Antimicrobial susceptibility testing
The antimicrobial susceptibility testing was carried out using the disc diffusion method on Mueller–Hinton agar (Oxoid, UK) according to the previous report (Wayne 2012). Erythromycin (15 μg), gentamicin (10 μg), tetracycline (30 μg), trimethoprim-sulphamethoxazole (25 μg), norfloxacin (10 μg), enrofloxacin (5 μg), streptomycin (10 μg), ampicillin (10 μg) and amoxicillin (20 μg) (Oxford, UK) were used. Results were estimated as described by the Clinical and Laboratory Standards Institute (CLSI 2016).
Statistical analysis
The Chi square test was used for the analysis of the obtained results using statistical analysis (SAS® software, version 9.4) to test the null hypothesis of various treatments.The significance level was (p < 0.05).
Results
Clinical examination
The clinical examination revealed pneumonia in calves approaching 68.4% (154/225), four calves died, and many cases showed mild calf scours. The farm record revealed respiratory infections, mortalities and scours a month prior to sample collection.
Prevalence of bacterial pathogens in apparently healthy and diseased calves
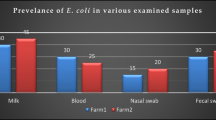
Bacterial pathogens were isolated from 57% of the examined calves with or without BRD (Table 2). The statistical analysis revealed that there is a significant difference in the prevalence of various pathogens among the examined apparently healthy and pneumonic calves (p < .05). The most prevalent bacterial pathogen isolated from calves with BRD was E. coli (23.5%), followed by P. vulgaris (12.4%) and S. aureus (11.8%). Moreover, E. coli was the most prevalent bacterium isolated from apparently healthy calves (16.4%), followed by S. aureus (10.9%) and P. vulgaris (5.4%). In pneumonic calves, P. multocida (n = 3) was isolated from lung tissue samples and M. haemolytica (n = 1) was isolated from blood samples (Table 3). No bacterial pathogens were detected in the blood samples that were collected from the apparently healthy calves. The bacterial examination yielded 128 isolates (109 from pneumonic calves and 19 from apparently healthy calves). Moreover, 17 samples (7.6%) presenting a combination of two bacterial pathogens (mixed infection) (Table 4).
Serotyping of the isolated E. coli isolates
The isolated E. coli (n = 49) strains were examined serologically with O and H antisera. As described in Table 5, the serological analysis of revealed 8 serogroups in pneumonic calves; O18:H6 (n = 5), O119:H4 (n = 4), O86:H9 (n = 5), O158:H10 (n = 4), O143:H4 (n = 8), O1:H2 (n = 4), O63:H7 (n = 6) and O128:H2 (n = 4), and 3 serogroups in apparently healthy calves; O18:H6 (n = 4), O86:H9 (n = 3), O63:H7 (n = 2). The statistical analysis proved that there is no significant difference in the types of the O antigens of E. coli isolated from healthy calves and pneumonic calves (p > 0.05).
Pathogenicity test for P. multocida isolates
All the isolates of P. multocida killed all the tested mice (n = 6) 24 h after the inoculation. The postmortem examination showed generalized septicemia with congestion at subcutaneous tissues and internal organs. The stained smears from heart blood showed the characteristic bipolar pattern of P. multocida.
PCR detection of virulence genes
As illustrated in Table 6, the fimH, eaeA and hly genes were detected in 100%, 24.5% and 20.4% of the isolated E. coli strains (n = 49), respectively. However, stx1 and stx2 genes were not detected in the tested E. coli strains. Furthermore, all the examined S. aureus strains (n = 26) harbored both spa and coa genes (100% each). Additionally, the toxA gene was detected in all the tested P. multocida strains.
Antimicrobial susceptibility testing
As illustrated in Tables 7 and 8, all the isolated E. coli strains were sensitive to enrofloxacin and norfloxacin (90% for each). Moreover, they exhibited multidrug-resistance against gentamicin (90%), erythromycin, streptomycin (80% for each) and trimethoprim-sulphamethoxazole (70%). Furthermore, the examined S. aureus strains showed remarkable sensitivity to enrofloxacin (90%), erythromycin, norfloxacin (80% for each) and gentamicin (70%), while showed multi-drug resistance pattern against amoxicillin, ampicillin (100% for each) and tetracycline (70%). In addition, all the examined P. aeruginosa strains were sensitive to enrofloxacin, norfloxacin and gentamicin, while showed multidrug-resistance against erythromycin, amoxicillin, ampicillin (100% for each) and tetracycline (80%). Regarding P. multocia, all the tested isolates were sensitive to gentamicin, enrofloxacin, and norfloxacin, while exhibited multidrug resistance against ampicillin, amoxicillin, erythromycin, and streptomycin (100% for each).
Discussion
This cross-sectional study analyzed the bacterial pathogens in healthy and pneumonic calves and provided a comprehensive analysis of the dominant bacterial pathogens causing respiratory infections in calves. The prevalence of causes of respiratory disease is highly changing within the cases and depends on various factors that affect the detected pathogens (Klima et al. 2014). Overall, the prevalence of bacterial pathogens in this study was 57%, comprising 62% of nasal swabs, 4.76% of blood samples and 75% of lung tissue (Table 2). These findings proved that the nasopharyngeal tract is a common habitat of opportunistic bacteria as well as the portal of entry of pathogenic infectious agents, which is endorsed by Holman et al. (Holman et al. 2015).
The most frequent bacterial pathogens isolated from the respiratory tract of apparently healthy calves were E. coli, followed by S. aureus, while in pneumonic calves were E. coli, P. vulgaris and S. aureus, these findings agree with previous studies characterizing the Proteobacteria in the nasopharyngeal tract of different animals (Holman et al. 2015; Ouchriah et al. 2015). Although de Oliveira et al. (de Oliveira et al. 2016) had isolated E. coli in a low proportion (4.22%) from healthy and diseased animals with respiratory signs, another report indicated the significance of E. coli in bronchopneumonia (DebRoy et al. 2008). The comparatively high proportion of E. coli in the study is worthwhile and of epidemiological importance. These findings suggest that these calves are likely to have originated from several farms and have a poor treatment and a negative immune deficiency and calls further characterization of more strains that will contribute to controlling the problem in local and/or international areas.
The bacterial variety of the bovine nasopharyngeal tract was much low when compared to other sites in the body (Holman et al. 2015). However, considerable different bacteria were observed in the present study from the nasal cavity of pneumonic calves greater than healthy calves as shown in Table 3, similar to previous reports (Angen et al. 2009; Clavijo et al. 2007; Ouchriah et al. 2015). Further, the finding of different bacterial pathogens in calves with pneumonia in this study supports the multifactorial causes of BRD in calves (Angen et al. 2009; Szeredi et al. 2010). A previous study revealed considerable variations in bacterial pathogens between healthy and diseased calves (Zeineldin et al. 2017).
In the current study, the E. coli serotyping revealed the serogroups; O86, O119, and O158, which were also demonstrated in diarrheic calves from Egypt by a previous report (Hakim et al. 2017). The serogroups; O18, O143, O1and O63 are specified in our survey and had a different variety than previous studies (Hakim et al. 2017; Vu-Khac and Cornick 2008), and to the best of our knowledge, this is the first report of these serogroups (O18, O143, O1, and O63) in E. coli strains identified from pneumonic calves in Egypt. Further, most E. coli strains isolated from healthy and pneumonic calves carry one to three virulence genes; fimH, eaeA and hly. However, stx1 and stx2 genes were not detected, in agreement with a previous study (Osman et al. 2013). Extra-intestinal E. coli carried many virulent genes that have been detected in bronchopneumonia in horse (DebRoy et al. 2008). These findings showed that all E. coli strains isolated in the current study from nasal secretions of pneumonic calves are being pathogenic and may play a role in the epidemiology of BRD either alone or in combination with other pathogens. Also, S. aureus is one of the most common identified bacterial pathogens isolated from healthy and pneumonic calves (Table 3). All examined S. aureus strains harbored both spa and coa virulence genes. The proportion of S. aureus isolation in this study was higher than reported in calves (1.4%) from Brazil (Gaeta et al. 2018). This variation may be due to the difference in hygienic measures, farm management, and exposure to stress. Calves can inhale many environmental bacteria and can detect those bacteria in both the upper and lower respiratory systems, and therefore their opportunistic role should be looked (Gaeta et al. 2018; Griffin et al. 2010).
Although P. multocida is a significant agent of BRD (Griffin et al. 2010), in the current study, lower prevalence (1.8%) of P. multocida was recorded in diseased calves when compared to a prior report (5.5%) described by de Oliveira et al. (de Oliveira et al. 2016). A previous study indicated that P. multocida is frequently isolated from calves with pneumonia, which has previously been exposed to environmental and stress factors (Dabo et al. 2007). In addition, the virulence of P. multocida was confirmed in this study, where all tested mice have died 24 h later, which agrees with the results obtained by Okerman et al. (Okerman et al. 1979). Further, the toxA gene is a virulence gene associated with P. multocida, was identified by PCR in all isolates. A previous investigation reported that PCR targeting toxA gene is sensitive and essential for the detection of pathogenic P. multocida (Varte et al. 2014).
Moreover, P. aeruginosa, E. cloacae, E. aerogenes, and M. haemolytica were detected (Table 3) more frequently from diseased calves compared to the healthy calves. Previous studies recovered Bacillus species, Klebsiella species and P. aeruginosa in calves with bronchopneumonia (de Oliveira et al. 2016). Regarding pathogens, some species that are not of obvious importance associated with pneumonia, but their opportunistic role should not be ignored (Griffin et al. 2010).
Regarding the antibiogram of the most predominant isolates, as shown in Tables 7 and 8, most of the isolated E. coli, S. aureus, P. multocida and P. aeruginosa strains were highly sensitive to enrofloxacin and norfloxacin and showed variable degrees of resistance against the rest of the tested antimicrobial agents. The high sensitivity of enrofloxacin and norfloxacin is supported by previous reports (Kroemer et al. 2012; Kumar et al. 2009) and suggests that these antibiotics can still be utilized with a high chance of curative success for the treatment of respiratory diseases in bovines. Furthermore, the finding of high effectiveness of gentamicin against P. multocida is in contrast with a previous study on veal calves (Catry et al. 2005), who found that P. multocida was gentamicin-resistant. The multidrug-resistance was observed among the majority of the isolated S. aureus, P. multocida and P. aeruginosa strains against 3-4 antimicrobial agents, while most of E. coli strains were resistant to 4 antimicrobial agents, and could be attributed to the random use of antibiotics over time in animal production (Haftu et al. 2012) and of clinical significance, particularly in the calves industry system. Previous studies from Ethiopia, Brazil, Belgium, Egypt, and Canada have indicated that there is a remarkable increase in the prevalence of multidrug-resistant E. coli, S. aureus and P. multocida strains (Catry et al. 2016; Eid et al. 2019; Enany et al. 2019; Haftu et al. 2012; Timsit et al. 2017, Algammal et al. 2020).
In conclusion, the current findings described the bacterial causes of BRD in nasal swabs, blood samples, and lung tissues obtained from apparently healthy and diseased calves. E. coli and S. aureus were the most predominant bacterial pathogens incriminated in BRD. The lower prevalence of important pathogens like P. multocida may indicate the possible role of other types of opportunistic bacteria in BRD. To the best of our knowledge, E. coli serogroups; O18, O143, O1, and O63 were identified in cases of calf pneumonia as the first report in Egypt. Continuous surveillance of antimicrobial susceptibility is essential to select the drug of choice due to the development of multidrug-resistant strains. Enrofloxacin and norfloxacin were the most effective antimicrobials against calf respiratory pathogens. The application of both phenotypic and genotypic analyses is more valuable as a diagnostic tool for identifying the causes and could help in the proper treatment of BRD.
Availability of data and materials
Not applicable.
References
Algammal AM, Enany ME, El-Tarabili RM, Ghobashy MOI, Helmy YA (2020) Prevalence, antimicrobial resistance profiles, virulence and enterotoxin-determinant genes of MRSA isolated from subclinical Bovine Mastitis in Egypt. Pathogens 9(5):E362. https://doi.org/10.3390/pathogens9050362
Angen Ø, Thomsen J, Larsen LE, Larsen J, Kokotovic B, Heegaard PM, Enemark JM (2009) Respiratory disease in calves: microbiological investigations on trans-tracheally aspirated bronchoalveolar fluid and acute phase protein response. Vet Microbiol 137:165–171
Bisi-Johnson MA, Obi CL, Vasaikar SD, Baba KA, Hattori T (2011) Molecular basis of virulence in clinical isolates of Escherichia coli and Salmonella species from a tertiary hospital in the Eastern Cape, South Africa. Gut Pathog 3:9
Catry B, Haesebrouck F, Vliegher SD, Feyen B, Vanrobaeys M, Opsomer G, Schwarz S, Kruif AD (2005) Variability in acquired resistance of Pasteurella and Mannheimia isolates from the nasopharynx of calves, with particular reference to different herd types. Microb drug resist 11:387–394
Catry B, Dewulf J, Maes D, Pardon B, Callens B, Vanrobaeys M, Opsomer G, de Kruif A, Haesebrouck F (2016) Effect of antimicrobial consumption and production type on antibacterial resistance in the bovine respiratory and digestive tract. PLoS ONE 11:e0146488
Clavijo A, Alfaro C, de Rolo M (2007) Ecología bacteriana relacionada con problemas respiratorios en terneros del estado Monagas. Zootec trop 25:197–200
CLSI (2016) Performance standards for antimicrobial susceptibility testing, vol 26. Clinical Lab Standards Institute, Wayne, PA, USA, pp 1–251
Dabo S, Taylor J, Confer A (2007) Pasteurella multocida and bovine respiratory disease. Anim Health Res Rev 8:129–150
de Oliveira BAFD, Gaeta NC, Ribeiro BLM, Alemán MAR, Marques LM, Timenetsky J, Melville PA, Marques JA, Marvulle V, Gregory L (2016) Determination of bacterial aetiologic factor on tracheobronchial lavage in relation to clinical signs of bovine respiratory disease. J Med Microbiol 65:1137–1142
DebRoy C, Roberts E, Jayarao BM, Brooks JW (2008) Bronchopneumonia associated with extraintestinal pathogenic Escherichia coli in a horse. J Vet Diagn Investig 20:661–664
Dipineto L, Santaniello A, Fontanella M, Lagos K, Fioretti A, Menna L (2006) Presence of Shiga toxin-producing Escherichia coli O157: H7 in living layer hens. Lett Appl Microbiol 43:293–295
Edwards PR, Ewing WH (1962) Identification of Enterobacteriaceae Identification of Enterobacteriaceae, 3rd Edition
Eid HI, Algammal AM, Nasef SA, Elfeil WK, Mansour GH (2016) Genetic variation among avian pathogenic E. coli strains isolated from broiler chickens. AJAVA 11:350–356
Eid HM, Algammal AM, Elfeil WK, Youssef FM, Harb SM, Abd-Allah EM (2019) Prevalence, molecular typing, and antimicrobial resistance of bacterial pathogens isolated from ducks. Vet World 12:677
Enany ME, Algammal AM, Shagar GI, Hanora AM, Elfeil WK, Elshaffy NM (2018) Molecular typing and evaluation of Sidr honey inhibitory effect on virulence genes of MRSA strains isolated from catfish in Egypt. Pak J Pharm Sci 31:1865
Enany ME, Algammal AM, Nasef SA, Abo-Eillil SA, Bin-Jumah M, Taha AE, Allam AA (2019) The occurrence of the multidrug resistance (MDR) and the prevalence of virulence genes and QACs resistance genes in E. coli isolated from environmental and avian sources. AMB Express 9:192
Gaeta NC, Ribeiro BL, Alemán MA, Yoshihara E, Nassar AF, Marques LM, Timenetsky J, Gregory L (2018) Bacterial pathogens of the lower respiratory tract of calves from Brazilian rural settlement herds and their association with clinical signs of bovine respiratory disease. Pesq Vet Bras 38:374–381
Ghanbarpour R, Salehi M (2010) Determination of adhesin encoding genes in Escherichia coli isolates from omphalitis of chicks. Am J Anim Vet Sci 5:91–96
Griffin D, Chengappa M, Kuszak J, McVey DS (2010) Bacterial pathogens of the bovine respiratory disease complex. Vet Clin N AM-Food A 26:381–394
Haftu R, Taddele H, Gugsa G, Kalayou S (2012) Prevalence, bacterial causes, and antimicrobial susceptibility profile of mastitis isolates from cows in large-scale dairy farms of Northern Ethiopia. Trop Anim Health Pro 44:1765–1771
Hakim AS, Omara ST, Syame SM, Fouad EA (2017) Serotyping, antibiotic susceptibility, and virulence genes screening of Escherichia coli isolates obtained from diarrheic buffalo calves in Egyptian farms. Vet World 10:769
Holman DB, Timsit E, Alexander TW (2015) The nasopharyngeal microbiota of feedlot cattle. Sci Rep 5:15557
Iyer AP, Kumosani TA PCR based detection of nosocomial infection causing MRSA (Methicillin resistant Staphylococcus aureus). In: 2nd International Conference on Biotechnology and Food Science IPCBEE, 2011
Klima CL, Zaheer R, Cook SR, Booker CW, Hendrick S, Alexander TW, McAllister TA (2014) Pathogens of bovine respiratory disease in North American feedlots conferring multidrug resistance via integrative conjugative elements. J Clin Microbiol 52:438–448
Kroemer S, Galland D, Guérin-Faublée V, Giboin H, Woehrlé-Fontaine F (2012) Survey of marbofloxacin susceptibility of bacteria isolated from cattle with respiratory disease and mastitis in Europe. Vet Rec 170:53
Kumar P, Singh V, Agrawal R, Singh S (2009) Identification of Pasteurella multocida isolates of ruminant origin using polymerase chain reaction and their antibiogram study. Trop Anim Health Pro 41:573–578
Okerman L, Spanoghe L, De Bruycker RM (1979) Experimental infections of mice with Pasteurella multocida strains isolated from rabbits. J Comp Pathol 89:51–55. https://doi.org/10.1016/0021-9975(79)90008-2
Osman K, Mustafa A, Elhariri M, Abdelhamed G (2013) The distribution of Escherichia coli serovars, virulence genes, gene association and combinations and virulence genes encoding serotypes in pathogenic E. coli recovered from diarrhoeic calves, sheep and goat. Transbound Emerg Dis 60:69–78
Ouchriah Y, Heleili N, Mamache B, Ayachi A, Kassah AL (2015) Antimicrobial sensitivity of bacterial strains isolated from new-born calves in the abattoir of Batna (Algeria). Int J Livest Res 5:32–42
Piva IC, Pereira AL, Ferraz LR, Silva RS, Vieira AC, Blanco JE, Blanco M, Blanco J, Giugliano LG (2003) Virulence markers of enteroaggregative Escherichia coli isolated from children and adults with diarrhea in Brasilia, Brazil. J Clin Microbiol 41:1827–1832
Quinn PJ, Markey BK, Leonard FC, Hartigan P, Fanning S, FitzPatrick E (2011) Veterinary microbiology and microbial disease. Wiley, Hoboken
Radostits OM, Gay CC, Hinchcliff KW, Constable PD (2006) Veterinary medicine e-book: a textbook of the diseases of cattle, horses, sheep, pigs and goats. Elsevier, Amsterdam
Szeredi L, Jánosi S, Pálfi V (2010) Microbiological and pathological examination of fatal calf pneumonia cases induced by bacterial and viral respiratory pathogens. Acta Vet Hung 58:341–356
Tang X, Zhao Z, Hu J, Wu B, Cai X, He Q, Chen H (2009) Isolation, antimicrobial resistance, and virulence genes of Pasteurella multocida strains from swine in China. J Clin Microbiol 47:951–958
Timsit E, Hallewell J, Booker C, Tison N, Amat S, Alexander TW (2017) Prevalence and antimicrobial susceptibility of Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni isolated from the lower respiratory tract of healthy feedlot cattle and those diagnosed with bovine respiratory disease. Vet Microbiol 208:118–125
Varte Z, Dutta T, Roychoudhury P, Begum J, Chandra R (2014) Isolation, identification, characterization and antibiogram of Pasteurella multocida isolated from pigs in Mizoram with special reference to progressive atrophic rhinitis. Vet World 7:95–99
Vu-Khac H, Cornick NA (2008) Prevalence and genetic profiles of Shiga toxin-producing Escherichia coli strains isolated from buffaloes, cattle, and goats in central Vietnam. Vet Microbiol 126:356–363
Wada M, Lkhagvadorj E, Bian L, Wang C, Chiba Y, Nagata S, Shimizu T, Yamashiro Y, Asahara T, Nomoto K (2010) Quantitative reverse transcription-PCR assay for the rapid detection of methicillin-resistant Staphylococcus aureus. J Appl Microbiol 108:779–788
Wayne P (2012) Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing Twenty-second informational supplement M100-S21 United States, Wayne
Zeineldin M, Lowe J, de Godoy M, Maradiaga N, Ramirez C, Ghanem M, El-Raof YA, Aldridge B (2017) Disparity in the nasopharyngeal microbiota between healthy cattle on feed, at entry processing and with respiratory disease. Vet Microbiol 208:30–37
Acknowledgements
The authors are grateful to the Researchers Supporting Project number (RSP-2019/53), King Saud University, Riyadh, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
AMA, MEE Designed the study; AMA, FMY, and SAS Methodology; AMA, MME, MOIG, GEB, and WNH Data analysis and statistical analysis; AMA and MME Writing, Review & Editing. All authors have agreed to the published version of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Handling of animals was conducted as described by the Animal Ethics Committee of Suez Canal University, Egypt.
Consent for publication
All authors gave their informed consent prior to their inclusion in the study.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Algammal, A.M., El-Sayed, M.E., Youssef, F.M. et al. Prevalence, the antibiogram and the frequency of virulence genes of the most predominant bacterial pathogens incriminated in calf pneumonia. AMB Expr 10, 99 (2020). https://doi.org/10.1186/s13568-020-01037-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13568-020-01037-z