Abstract
Key message
The present study provides comparative transcriptome analysis, besides identifying functional secondary metabolite genes of Plumbago zeylanica with pharmacological potential for future functional genomics, and metabolomic engineering of secondary metabolites from this plant towards diversified biomedical applications.
Abstract
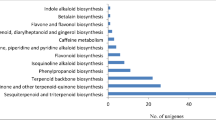
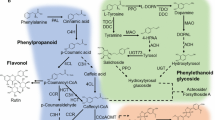
Plumbago zeylanica is a widely used medicinal plant of the traditional Indian system of medicine with wide pharmacological potential to treat several disorders. The present study aimed to carry out comparative transcriptome analysis in leaf and root tissue of P. zeylanica using Illumina paired end sequencing to identify tissue-specific functional genes involved in the biosynthesis of secondary metabolites, contributing to its therapeutic efficacy. De novo sequencing assembly resulted in the identification of 62,321 “Unigenes” transcripts with an average size of 1325 bp. Functional annotation using BLAST2GO resulted in the identification of 50,301 annotated transcripts (80.71%) and GO assigned to 18,814 transcripts. KEGG pathway annotation of the “Unigenes” revealed that 2465 transcripts could be assigned to 242 KEGG pathway maps wherein the number of transcripts involved in secondary metabolism was distinct in root and leaf transcriptome. Among the secondary metabolite biosynthesis pathways, the cluster of “Unigenes” encoding enzymes of ‘Phenylpropanoid biosynthesis pathway’ represents the largest group (84 transcripts) followed by ‘Terpenoid Backbone biosynthesis’ (48 transcripts). The transcript levels of the candidate unigenes encoding key enzymes of phenylpropanoid (PAL, TAL) and flavanoid biosynthesis (CHS, ANS, FLS) pathways were up-regulated in root, while the expression levels of candidate “Unigenes” transcript for monoterpenoid (DXS, ISPF), diterpenoid biosynthesis (SPS, SDS) and indole alkaloid pathways (STR) were significantly higher in leaf of P. zeylanica. Interestingly, validation of differential gene expression profile by qRT-PCR also confirmed that candidate “Unigenes” enzymes of phenylpropanoid and flavonoid biosynthesis were highly expressed in the root, while the key regulatory enzymes of terpenoid and indole alkaloid compounds were up-regulated in the leaf, suggesting that (differences in) the levels of these functional genes could be attributed to the (differential) pharmacological activity (between root and leaf) in tissues of P. zeylanica.







Similar content being viewed by others
References
Adisakwattana S (2017) Cinnamic acid and its derivatives: mechanisms for prevention and management of diabetes and its complications. Nutrients 9:163
Anandan A, Eswaran R, Doss A, Sangeetha G, Anand SP (2012) Chemical compounds investigation of Lucas Aspera leaves—a potential folklore medicinal plant. Asian J Pharm Clin Res 5:86–88
Annadurai RS, Neethiraj R, Jayakumar V, Damodaran AC, Rao SN, Katta MA, Gopinathan S, Sarma SP, Senthilkumar V, Niranjan V (2013) De novo transcriptome assembly (NGS) of Curcuma longa L. rhizome reveals novel transcripts related to anticancer and antimalarial terpenoids. PLoS ONE 8:56217. https://doi.org/10.1371/journal.pone.0056217
Bankar KG, Todur VN, Shukla RN, Vasudevan M (2015) Ameliorated de novo transcriptome assembly using Illumina paired end sequence data with trinity assembler. Genom Data 5:352–359
Bowles DJ, Isayenkova J, Lim EK, Poppenberger B (2005) Glycosyltransferases: managers of small molecules. Curr Opin Plant Biol 8:254–263
Camon E, Magrane M, Barrell D, Lee V, Dimmer E, Maslen J, Binns D, Harte N, Lopez R, Apweiler R (2004) The gene ontology annotation (GOA) database: sharing knowledge in uniprot with gene ontology. Nucleic Acids Res 32:262–266
Carson CF, Hammer KA (2010) Chemistry and bioactivity of essential oils. In: Thormar H (ed) Lipids and essential oils as antimicrobial agents. John Wiley and Sons, New York, pp 203–238
Chauhan M (2014) A review on morphology, phytochemistry and pharmacological activities of medicinal herb Plumbago Zeylanica Linn. J Pharmacogn Phytochem 3(2):95–118
Chen C, Li A (2016) Transcriptome analysis of differentially expressed genes involved in proanthocyanidin accumulation in the rhizomes of Fagopyrum dibotrys and an irradiation-induced mutant. Front Physiol 7:100. https://doi.org/10.3389/fphys.2016.00100
Chhetri SBB, Khatri D (2017) Phytochemical screening, total phenolic and flavonoid content and antioxidant activity of selected nepalese plants. World J Pharm Pharm Sci 6:951–968
Cuong DM, Kwon SJ, Jeon J, Park YJ, Park JS, Park SU (2018) Identification and characterization of phenyl propanoid biosynthetic genes and their accumulation in bitter melon (Momordica charantia). Molecules 23:469. https://doi.org/10.3390/molecules23020469
Dao TTH, Linthorst HJM, Verpoorte R (2011) Chalcone synthase and its functions in plant resistance. Phytochem Rev 10:397–412
de Hoon MJ, Imoto S, Nolan J, Miyano S (2004) Open source clustering software. Bioinformatics 20:1453–1454
Ferreyra MLF, Rius SP, Casati P (2012) Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front Plant Sci 3:222. https://doi.org/10.3389/fpls.2012.00222
Fukushima A, Nakamura M, Suzuki H, Yamazaki M, Knoch E, Mori T, Umemoto N, Morita M, Hirai G, Sodeoka M, Saito K (2016) Comparative characterization of the leaf tissue of Physalis alkekengi and Physalis peruviana using RNA-seq and metabolite profiling. Front Plant Sci 7:1883. https://doi.org/10.3389/fpls.2016.01883
Ganesan K, Gani SB (2013) Ethnomedical and pharmacological potentials of Plumbago zeylanica L—a review. Am J Phytomed Clin Ther 1(3):313–337
Guo X, Li Y, Li C, Luo H, Wang L, Qian J, Luo X, Xiang L, Song J, Sun C, Xu H, Yao H, Chen S (2013) Analysis of the Dendrobium officinale transcriptome reveals putative alkaloid biosynthetic genes and genetic markers. Gene 527:131–138
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J et al (2013) De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512
Hemaiswarya S, Doble M (2013) Combination of phenyl propanoids with 5-fluorouracil as anti-cancer agents against human cervical cancer (HeLa) cell line. Phytomedicine 20:151–158
Hiradeve S, Danao K, Kharabe V, Mendhe B (2011) Evaluation of anticancer activity of Plumbago zeylanica Linn. leaf extract. Int J Biomed Sci 1:01–09
Ittiyavirah SP, Ruby R (2014) Effect of hydro-alcoholic root extract of Plumbago zeylanica alone and its combination with aqueous leaf extract of Camellia sinensis on haloperidol induced parkinsonism in Wistar rats. Ann Neurosci 21:47–50
Jadhav SKR, Patel KA, Dholakia BB, Khan BM (2012) Structural characterization of a flavonoid glycosyltransferase from Withania somnifera. Bioinformation 8:943–949
Jain P, Sharma HP, Basri F, Baraik B, Kumari S, Pathak C (2014) Pharmacological profiles of ethno-medicinal plant: Plumbago zeylanica l.—a review. Int J Pharm Sci Rev Res 24:157–163
Jeena GS, Fatima S, Tripathi P, Upadhyay S, Shukla RK (2017) Comparative transcriptome analysis of shoot and root tissue of Bacopa monnieri identifies potential genes related to triterpenoid saponin biosynthesis. BMC Genom 18:490. https://doi.org/10.1186/s12864-017-3865-5
Ji Y, Xiao J, Shen Y, Ma D, Li Z, Pu G, Li X, Huang L, Liu B, Ye H (2014) Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in Artemisia annua. Plant Cell Physiol 55:1592–1604
Jiang N, Doseff AI, Grotewold E (2016) Flavones: from biosynthesis to health benefits. Plants 5:27. https://doi.org/10.3390/plants5020027
Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45:353–361
Kawiak A, Domachowska A, Jaworska A, Lojkowska E (2017) Plumbagin sensitizes breast cancer cells to tamoxifen-induced cell death through GRP78 inhibition and Bik upregulation. Sci Rep. https://doi.org/10.1038/srep43781
Kumar S, Pandey AK (2013) Chemistry and biological activities of flavonoids: an overview. Sci World J. https://doi.org/10.1155/2013/162750
Kumar A, Kumar S, Bains S, Vaidya V, Singh B, Kaur R, Kaur J, Singh K (2016) De novo transcriptome analysis revealed genes involved in flavonoid and vitamin C biosynthesis in Phyllanthus emblica (L). Front Plant Sci 7:1610. https://doi.org/10.3389/fpls.2016.01610
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform 12:323
Lin D, Xiao M, Zhao J, Li Z, Xing B, Li X, Kong M, Li L, Zhang Q, Liu Y, Chen H, Qin W, Wu H, Chen S (2016) An overview of plant phenolic compounds and their importance in human nutrition and management of type 2 diabetes. Molecules 21:1374. https://doi.org/10.3390/molecules21101374
Liu Y, Zhu X, Li W, Wen H, Gao Y, Liu Y, Liu C (2014) Enhancing production of ergosterol in Pichia pastoris GS115 by over-expression of 3-hydroxy-3-methylglutaryl CoA reductase from Glycyrrhiza uralensis. Acta Pharm Sin B 4:161–166
Liu Y, Wang Y, Guo F, Zhan L, Mohr T, Cheng P, Huo N, Gu R, Pei D, Sun J, Tang L, Long C, Huang L, Gu YQ (2017) Deep sequencing and transcriptome analyses to identify genes involved in secoiridoid biosynthesis in the Tibetan medicinal plant Swertia mussotii. Sci Rep 7:43108. https://doi.org/10.1038/srep43108
Liu QQ, Luo L, Zheng L (2018) Lignins: biosynthesis and biological functions in plants. Int J Mol Sci. https://doi.org/10.3390/ijms19020335
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25:402–408
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550
Lu X, Tang K, Li P (2016) Plant metabolic engineering strategies for the production of pharmaceutical terpenoids. Front Plant Sci. https://doi.org/10.3389/fpls.2016.01647
Malar RJJT (2013) Studies on interspecific and intraspecific variation in the genus: Plumbago (Plumbaginaceae) from south India. Dissertation, Manonmaniam Sundaranar University, Tamil Nadu.
Mandavkar YD, Jalalpure SS (2011) A comprehensive review on Plumbago zeylanica Linn. Afr J Pharm Pharmacol 5:2738–2747
Mazumdar AB, Chattopadhyay S (2015) Sequencing, de novo assembly, functional annotation and analysis of Phyllanthus amarus leaf transcriptome using the Illumina platform s. Front Plant Sci 6:1199. https://doi.org/10.3389/fpls.2015.01199
Miralpeix B, Sabalza M, Twyman RM, Capell T, Christou P (2014) Strategic patent analysis in plant biotechnology: terpenoid indole alkaloid metabolic engineering as a case study. Plant Biotechnol J 12:117–134
Misra BB (2014) An updated snapshot of recent advances in transcriptomics and genomics of phytomedicinals. J Postdoct Res 2:1–15
Ndikuryayo F, Moosavi B, Yang W-C, Yang G-F (2017) 4-hydroxyphenyl pyruvate dioxygenase inhibitors: from chemical biology to agrochemicals. J Agric Food Chem 65:8523–8537
Nile SH, Khobragade CN (2010) Antioxidant activity and flavonoid derivatives of Plumbago zeylanica. J Nat Prod 3:130–133
Pastore S, Mariani V, Cesareo E, Korkina L (2007) Anti-inflammatory effects of phenyl propanoids on human keratinocytes. Free Radic Res 41:46
Raghu D, Karunagaran D (2014) Plumbagin down regulates Wnt signaling independent of p53 in human colorectal cancer cells. J Nat Prod 77:1130–1134
Rahnamaie-Tajadod R, Loke KK, Goh HH, Noor NM (2017) Differential gene expression analysis in Polygonum minus leaf upon 24 h of methyl jasmonate elicitation. Front Plant Sci. https://doi.org/10.3389/fpls.2017.00109
Rajakrishnan R, Lekshmi R, Benil PB, Thomas J, AlFarhan AH, Rakesh V, Khalaf S (2017) Phytochemical evaluation of roots of Plumbago zeylanica L. and assessment of its potential as a nephroprotective agent. Saudi J Biol Sci 24:760–766
Rios JL (2010) Effects of triterpenes on the immune system. J Ethnopharmacol 128:1–14
Rondeau G, Abedinpour P, Chrastina A, Pelayo J, Borgstrom P, Welsh J (2018) Differential gene expression induced by anti-cancer agent plumbagin is mediated by androgen receptor in prostate cancer cells. Sci Rep 8:2694. https://doi.org/10.1038/s41598-018-20451-9
Roy A, Bharadvaja N (2017) A review on pharmaceutically important medical plant: Plumbago zeylanica. J Ayu Herb Med 3:225–228
Roy A, Attre T, Bharadvaja N (2017) Anticancer agent from medicinal plants: a review. In: Tiezzi A (ed) New aspects in medicinal plants and pharmacognosy. JB Books Publisher, Poland
Ru M, Wang K, Bai Z, Peng L, He S, Wang Y, Liang Z (2017) A tyrosine aminotransferase involved in rosmarinic acid biosynthesis in Prunella vulgaris L. Sci Rep. https://doi.org/10.1038/s41598-017-05290-4
Saldanha AJ (2004) Java Treeview—extensible visualization of microarray data. Bioinformatics 20:3246–3248
Satyavati VG, Gupta KA (1987) Medicinal plants of India. Indian Council Med Res 2:471–477
Savoia D (2012) Plant-derived antimicrobial compounds: alternatives to antibiotics. Fut Microbiol 7:979–990
Schluttenhofer C, Yuan L (2015) Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. https://doi.org/10.1104/pp.114.251769
Shivashankar S, Sumathi M, Krishnakumar NK, Rao VK (2015) Role of phenolic acids and enzymes of phenylpropanoid pathway in resistance of chayote fruit (Sechium edule) against infestation by melon fly Bactrocera cucurbitae. Ann Appl Biol 166:420–433
Shukla AK, Mall M, Rai SK, Singh S, Nair P, Parashar G, Shasany AK, Singh SC, Joshi VK, Khanuja SPS (2013) A transcriptomic approach for exploring the molecular basis for dosha-balancing property-based classification of plants in Ayurveda. Mol Biol Rep 40:3255–3262
Singh J, Mishra NP, Jain SP, Singh SC, Sharma A, Khanuja SPS (2004) Traditional uses of Plumbago zeylanica (Chitrak). J Med Arom Plant Sci 26:795–800
Singh M, Nagori K, Iyer S, Khare G, Sharwan G, Tripathi DK (2011) Ethano-medicinal, traditional and pharmacological aspects of Plumbago zeylanica. Pharmacol Online Newslett 3:684–700
Singh S, Sharma B, Kanwar SS, Kumar A (2016) Lead phytochemicals for anticancer drug development. Front Plant Sci 7:1667. https://doi.org/10.3389/fpls.2016.01667
Srivastava S, Sanchita SR, Srivastava G, Sharma A (2018) Comparative study of withanolide biosynthesis-related miRNAs in root and leaf tissues of Withania somnifera. Appl Biochem Biotechnol. https://doi.org/10.1007/s12010-018-2702-x
Stewart CJR, Vickery CR, Burkart MD, Noel JP (2013) Confluence of structural and chemical biology: plant polyketide synthases as biocatalysts for a bio-based future. Curr Opin Plant Biol 16:365–372
Stockigta J, Barleben L, Panjikar S, Lorisa EA (2008) 3D-Structure and function of strictosidine synthase—the key enzyme of monoterpenoid indole alkaloid biosynthesis. Plant Physiol Biochem 46:340–355
Stracke R, Ishihara H, Barsch GHA, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. The Plant J 50:660–677
Sundari BKR, Telapolu S, Dwarakanath BS, Thyagarajan SP (2017) Cytotoxic and antioxidant effects in various tissue extracts of Plumbago zeylanica: implications for anticancer potential. Phcog J 9:706–712
Sunil C, Duraipandiyan V, Agastian P, Ignacimuthu S (2012) Antidiabetic effect of plumbagin isolated from Plumbago zeylanica L. root and its effect on GLUT4 translocation in streptozotocin-induced diabetic rats. Food Chem Toxicol 50:4356–4363
Suttipanta N, Pattanaik S, Kulshrestha M, Patra B, Singh SK, Yuan L (2011) The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol 157:2081–2093
Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422
Tholl D (2006) Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr Opin Plant Biol 9:297–304
Tholl D (2015) Biosynthesis and biological functions of terpenoids in plants. Adv Biochem Eng Biotechnol 148:63–106
Upadhyay S, Phukan UJ, Mishra S, Shukla RK (2014) De novo leaf and root transcriptome analysis identified novel genes involved in Steroidal sapogenin biosynthesis in Asparagus racemosus. BMC Genomics 15:746
Vishnukanta RAC (2008) Analgesic and anti-inflammatory activity of hydroalcoholic extract of Plumbago zeylanica leaf extract. Pharmacogn Mag 15:S133–136
Widhalm JR, Rhodes D (2016) Biosynthesis and molecular actions of specialized 1,4-naphthoquinone natural products produced by horticultural plants. Hortic Res. https://doi.org/10.1038/hortres.2016.46
Yang H, Dou QP (2010) Targeting apoptosis pathway with natural terpenoids: implications for treatment of breast and prostate cancer. Curr Drug Targets 11:733–744
Yang L, Ding G, Lin H, Cheng H, Kong Y, Wei Y, Fang X, Liu R, Wang L, Chen X, Yang C (2013a) Transcriptome analysis of medicinal plant salvia miltiorrhiza and identification of genes related to tanshinone biosynthesis. PLoS ONE 8:e80464. https://doi.org/10.1371/journal.pone.0080464
Yang L, Ding G, Lin H, Cheng H, Kong Y, Wei Y, Fang X, Liu R, Wang L, Chen X, Yang C (2013b) Transcriptome analysis of medicinal plant Salvia miltiorrhiza and identification of genes related to tanshinone biosynthesis. PLoS ONE 8:e80464. https://doi.org/10.1371/journal.pone.0080464
Zhang H, Hedhili S, Montiel G, Zhang Y, Chatel G, Pre M, Gantet P, Memelink J (2011) The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J 67:61–71
Zhang K, Chen D, Ma K, Wu X, Hao H, Jiang S (2018) NAD(P)H: quinone oxidoreductase 1 (NQO1) as a therapeutic and diagnostic target in cancer. J Med Chem. https://doi.org/10.1021/acs.jmedchem.8b00124
Zhu W, Yang B, Komatsu S, Lu X, Li X, Tian J (2015) Binary stress induces an increase in indole alkaloid biosynthesis in Catharanthus roseus. Front. Plant Sci. https://doi.org/10.3389/fpls.2015.00582
Acknowledgements
The author Karpaga Raja Sundari acknowledges DST-National Post-Doctoral Fellowship for funding this research work. KRS also acknowledges the support of Dr. Modhumita Dasgupta, Scientist F, Institute of Forest Genetics and Tree Breeding, Coimbatore, for providing the guidance and facilities to carry out RNA extraction from tissues of P. zeylanica. The study was funded by Science and Engineering Research Board(SERB) (Grant no. DST/NPDF/2015/000524).
Author information
Authors and Affiliations
Contributions
The author BKRS and SPT conceptualized and designed the experiments for RNA sequencing. RB contributed to RNA sequencing and acquisition of data while BKRS and RB analyzed the transcriptome data. BKRS conducted experimental validation by qRT-PCR for the obtained transcripts. BKRS and BSD interpreted the data and drafted the article. BSD and SPT critically revised the article and made the final approval of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The corresponding author, Karpaga Raja Sundari has received grant from Department of science and Technology, Govt. of India. All the authors declare that they have no competing financial and non-financial interests.
Data archiving statement
P. zeylanica leaf and root transcriptome raw data can be accessed at NCBI SRA database under SRA ID SRP144988.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Karpaga Raja Sundari, B., Budhwar, R., Dwarakanath, B.S. et al. De novo transcriptome analysis unravels tissue-specific expression of candidate genes involved in major secondary metabolite biosynthetic pathways of Plumbago zeylanica: implication for pharmacological potential. 3 Biotech 10, 271 (2020). https://doi.org/10.1007/s13205-020-02263-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02263-9