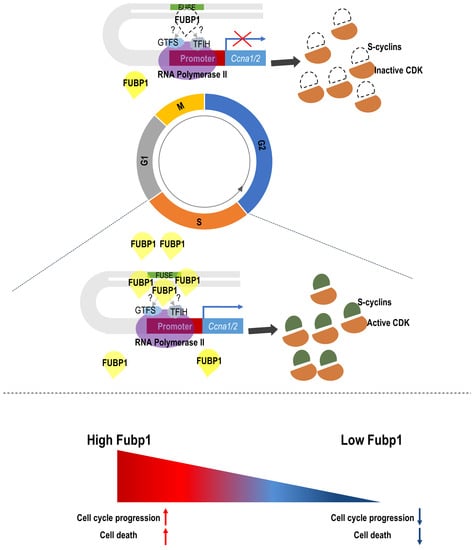
Multiple Functions of Fubp1 in Cell Cycle Progression and Cell Survival
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. Cell Culture and Generation of Fubp1-Deficient LLC Cells
2.3. Protein Preparation and Immunoblot Analysis
2.4. Immunofluorescence
2.5. Quantitative Real-Time RT-PCR
2.6. Chromatin Immunoprecipitation (ChIP) Assay
2.7. Patients and Samples
2.8. Statistical Analysis
3. Results
3.1. Cyclin A Is a Transcriptional Target of Fubp1
3.2. Fubp1 Deficiency Slows Down Cell Cycle Progression
3.3. Fubp1 Expression Patterns Show Heterogeneity among Cancer Tissues
3.4. Fubp1 Is Involved in Both Cell Cycling and Cell Survival
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Avigan, M.I.; Strober, B.; Levens, D. A far upstream element stimulates c-myc expression in undifferentiated leukemia cells. J. Biol. Chem. 1990, 265, 18538–18545. [Google Scholar] [PubMed]
- He, L.; Weber, A.; Levens, D. Nuclear targeting determinants of the far upstream element binding protein, a c-myc transcription factor. Nucleic Acids Res. 2000, 28, 4558–4565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olanich, M.E.; Moss, B.L.; Piwnica-Worms, D.; Townsend, R.R.; Weber, J.D. Identification of FUSE-binding protein 1 as a regulatory mRNA-binding protein that represses nucleophosmin translation. Oncogene 2011, 30, 77–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chien, H.L.; Liao, C.L.; Lin, Y.L. FUSE binding protein 1 interacts with untranslated regions of Japanese encephalitis virus RNA and negatively regulates viral replication. J. Virol. 2011, 85, 4698–4706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malz, M.; Weber, A.; Singer, S.; Riehmer, V.; Bissinger, M.; Riener, M.O.; Longerich, T.; Soll, C.; Vogel, A.; Angel, P.; et al. Overexpression of far upstream element binding proteins: A mechanism regulating proliferation and migration in liver cancer cells. Hepatology 2009, 50, 1130–1139. [Google Scholar] [CrossRef]
- Rabenhorst, U.; Beinoraviciute-Kellner, R.; Brezniceanu, M.L.; Joos, S.; Devens, F.; Lichter, P.; Rieker, R.J.; Trojan, J.; Chung, H.J.; Levens, D.L.; et al. Overexpression of the far upstream element binding protein 1 in hepatocellular carcinoma is required for tumor growth. Hepatology 2009, 50, 1121–1129. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.H.; Hu, J.L.; Liang, J.Z.; Zhou, A.J.; Li, M.Z.; Yan, S.M.; Zhang, X.; Gao, S.; Chen, L.; Zhong, Q.; et al. Far upstream element-binding protein 1 is a prognostic biomarker and promotes nasopharyngeal carcinoma progression. Cell Death Dis. 2015, 6, e1920. [Google Scholar] [CrossRef] [Green Version]
- Venturutti, L.; Cordo Russo, R.I.; Rivas, M.A.; Mercogliano, M.F.; Izzo, F.; Oakley, R.H.; Pereyra, M.G.; De Martino, M.; Proietti, C.J.; Yankilevich, P.; et al. MiR-16 mediates trastuzumab and lapatinib response in ErbB-2-positive breast and gastric cancer via its novel targets CCNJ and FUBP1. Oncogene 2016, 35, 6189–6202. [Google Scholar] [CrossRef]
- Hoang, V.T.; Verma, D.; Godavarthy, P.S.; Llavona, P.; Steiner, M.; Gerlach, K.; Michels, B.E.; Bohnenberger, H.; Wachter, A.; Oellerich, T.; et al. The transcriptional regulator FUBP1 influences disease outcome in murine and human myeloid leukemia. Leukemia 2019, 33, 1700–1712. [Google Scholar] [CrossRef]
- Jiang, P.; Huang, M.; Qi, W.; Wang, F.; Yang, T.; Gao, T.; Luo, C.; Deng, J.; Yang, Z.; Zhou, T.; et al. FUBP1 promotes neuroblastoma proliferation via enhancing glycolysis-a new possible marker of malignancy for neuroblastoma. J. Exp. Clin. Cancer Res. 2019, 38, 400. [Google Scholar] [CrossRef]
- Ding, Z.; Liu, X.; Liu, Y.; Zhang, J.; Huang, X.; Yang, X.; Yao, L.; Cui, G.; Wang, D. Expression of far upstream element (FUSE) binding protein 1 in human glioma is correlated with c-Myc and cell proliferation. Mol. Carcinog 2015, 54, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Atanassov, B.S.; Dent, S.Y. USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1. EMBO Rep. 2011, 12, 924–930. [Google Scholar] [CrossRef]
- Weber, A.; Kristiansen, I.; Johannsen, M.; Oelrich, B.; Scholmann, K.; Gunia, S.; May, M.; Meyer, H.A.; Behnke, S.; Moch, H.; et al. The FUSE binding proteins FBP1 and FBP3 are potential c-myc regulators in renal, but not in prostate and bladder cancer. BMC Cancer 2008, 8, 369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, J.; Bao, X.; Ma, X.; Zhang, Y.; Ni, D.; Wang, H.; Zhang, F.; Du, Q.; Fan, Y.; Chen, J.; et al. Upregulation of Far Upstream Element-Binding Protein 1 (FUBP1) Promotes Tumor Proliferation and Tumorigenesis of Clear Cell Renal Cell Carcinoma. PLoS ONE 2017, 12, e0169852. [Google Scholar] [CrossRef] [PubMed]
- Debaize, L.; Jakobczyk, H.; Avner, S.; Gaudichon, J.; Rio, A.G.; Serandour, A.A.; Dorsheimer, L.; Chalmel, F.; Carroll, J.S.; Zornig, M.; et al. Interplay between transcription regulators RUNX1 and FUBP1 activates an enhancer of the oncogene c-KIT and amplifies cell proliferation. Nucleic Acids Res. 2018, 46, 11214–11228. [Google Scholar] [CrossRef] [Green Version]
- Bettegowda, C.; Agrawal, N.; Jiao, Y.; Sausen, M.; Wood, L.D.; Hruban, R.H.; Rodriguez, F.J.; Cahill, D.P.; McLendon, R.; Riggins, G.; et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science 2011, 333, 1453–1455. [Google Scholar] [CrossRef] [Green Version]
- Hwang, I.; Cao, D.; Na, Y.; Kim, D.Y.; Zhang, T.; Yao, J.; Oh, H.; Hu, J.; Zheng, H.; Yao, Y.; et al. Far Upstream Element-Binding Protein 1 Regulates LSD1 Alternative Splicing to Promote Terminal Differentiation of Neural Progenitors. Stem Cell Rep. 2018, 10, 1208–1221. [Google Scholar] [CrossRef] [Green Version]
- Molenaar, J.J.; Koster, J.; Zwijnenburg, D.A.; van Sluis, P.; Valentijn, L.J.; van der Ploeg, I.; Hamdi, M.; van Nes, J.; Westerman, B.A.; van Arkel, J.; et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 2012, 483, 589–593. [Google Scholar] [CrossRef]
- Girard, F.; Strausfeld, U.; Fernandez, A.; Lamb, N.J. Cyclin A is required for the onset of DNA replication in mammalian fibroblasts. Cell 1991, 67, 1169–1179. [Google Scholar] [CrossRef]
- Fung, T.K.; Siu, W.Y.; Yam, C.H.; Lau, A.; Poon, R.Y. Cyclin F is degraded during G2-M by mechanisms fundamentally different from other cyclins. J. Biol. Chem. 2002, 277, 35140–35149. [Google Scholar] [CrossRef] [Green Version]
- Koff, A.; Giordano, A.; Desai, D.; Yamashita, K.; Harper, J.W.; Elledge, S.; Nishimoto, T.; Morgan, D.O.; Franza, B.R.; Roberts, J.M. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science 1992, 257, 1689–1694. [Google Scholar] [CrossRef] [PubMed]
- Dickson, C.; Fantl, V.; Gillett, C.; Brookes, S.; Bartek, J.; Smith, R.; Fisher, C.; Barnes, D.; Peters, G. Amplification of chromosome band 11q13 and a role for cyclin D1 in human breast cancer. Cancer Lett. 1995, 90, 43–50. [Google Scholar] [CrossRef]
- Gillett, C.; Fantl, V.; Smith, R.; Fisher, C.; Bartek, J.; Dickson, C.; Barnes, D.; Peters, G. Amplification and overexpression of cyclin D1 in breast cancer detected by immunohistochemical staining. Cancer Res. 1994, 54, 1812–1817. [Google Scholar] [PubMed]
- Husdal, A.; Bukholm, G.; Bukholm, I.R. The prognostic value and overexpression of cyclin A is correlated with gene amplification of both cyclin A and cyclin E in breast cancer patient. Cell Oncol. 2006, 28, 107–116. [Google Scholar] [PubMed]
- Buckley, M.F.; Sweeney, K.J.; Hamilton, J.A.; Sini, R.L.; Manning, D.L.; Nicholson, R.I.; deFazio, A.; Watts, C.K.; Musgrove, E.A.; Sutherland, R.L. Expression and amplification of cyclin genes in human breast cancer. Oncogene 1993, 8, 2127–2133. [Google Scholar]
- Platzer, P.; Upender, M.B.; Wilson, K.; Willis, J.; Lutterbaugh, J.; Nosrati, A.; Willson, J.K.; Mack, D.; Ried, T.; Markowitz, S. Silence of chromosomal amplifications in colon cancer. Cancer Res. 2002, 62, 1134–1138. [Google Scholar]
- Sanjana, N.E.; Shalem, O.; Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 2014, 11, 783–784. [Google Scholar] [CrossRef] [Green Version]
- Chung, H.J.; Liu, J.; Dundr, M.; Nie, Z.; Sanford, S.; Levens, D. FBPs are calibrated molecular tools to adjust gene expression. Mol. Cell Biol. 2006, 26, 6584–6597. [Google Scholar] [CrossRef] [Green Version]
- Kang, M.; Lee, S.M.; Kim, W.; Lee, K.H.; Kim, D.Y. Fubp1 supports the lactate-Akt-mTOR axis through the upregulation of Hk1 and Hk2. Biochem. Biophys. Res. Commun. 2019, 512, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, L.R.; Chung, H.J.; Sanford, S.; Kouzine, F.; Liu, J.; Levens, D. Hierarchical mechanisms build the DNA-binding specificity of FUSE binding protein. Proc. Natl. Acad. Sci. USA 2008, 105, 18296–18301. [Google Scholar] [CrossRef] [Green Version]
- Fotedar, A.; Cannella, D.; Fitzgerald, P.; Rousselle, T.; Gupta, S.; Doree, M.; Fotedar, R. Role for cyclin A-dependent kinase in DNA replication in human S phase cell extracts. J. Biol. Chem. 1996, 271, 31627–31637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horie, K.; Yamamoto, H.; Karube, K.; Takebayashi, K.; Yoshino, H.; Yoshioka, H.; Watanabe, J. Cyclin A is a reliable proliferation marker in endometrial cancer cell lines. Oncol. Lett. 2019, 17, 4455–4462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, L.; Zhai, L.; Qian, L.; Huang, D.; Ding, Y.; Xiang, H.; Liu, X.; Thompson, J.W.; Liu, J.; He, Y.H.; et al. Switching off IMMP2L signaling drives senescence via simultaneous metabolic alteration and blockage of cell death. Cell Res. 2018, 28, 625–643. [Google Scholar] [CrossRef] [Green Version]
- Oda, T.; Sekimoto, T.; Kurashima, K.; Fujimoto, M.; Nakai, A.; Yamashita, T. Acute HSF1 depletion induces cellular senescence through the MDM2-p53-p21 pathway in human diploid fibroblasts. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef] [Green Version]
- Reifenberger, G.; Weber, R.G.; Riehmer, V.; Kaulich, K.; Willscher, E.; Wirth, H.; Gietzelt, J.; Hentschel, B.; Westphal, M.; Simon, M.; et al. Molecular characterization of long-term survivors of glioblastoma using genome- and transcriptome-wide profiling. Int. J. Cancer 2014, 135, 1822–1831. [Google Scholar] [CrossRef] [PubMed]
- Oghumu, S.; Knobloch, T.J.; Terrazas, C.; Varikuti, S.; Ahn-Jarvis, J.; Bollinger, C.E.; Iwenofu, H.; Weghorst, C.M.; Satoskar, A.R. Deletion of macrophage migration inhibitory factor inhibits murine oral carcinogenesis: Potential role for chronic pro-inflammatory immune mediators. Int. J. Cancer 2016, 139, 1379–1390. [Google Scholar] [CrossRef] [Green Version]
- Ming, X.Y.; Zhang, X.; Cao, T.T.; Zhang, L.Y.; Qi, J.L.; Kam, N.W.; Tang, X.M.; Cui, Y.Z.; Zhang, B.Z.; Li, Y.; et al. RHCG Suppresses Tumorigenicity and Metastasis in Esophageal Squamous Cell Carcinoma via Inhibiting NF-kappaB Signaling and MMP1 Expression. Theranostics 2018, 8, 185–198. [Google Scholar] [CrossRef]
- Zhang, G.; He, P.; Tan, H.; Budhu, A.; Gaedcke, J.; Ghadimi, B.M.; Ried, T.; Yfantis, H.G.; Lee, D.H.; Maitra, A.; et al. Integration of metabolomics and transcriptomics revealed a fatty acid network exerting growth inhibitory effects in human pancreatic cancer. Clin. Cancer Res. 2013, 19, 4983–4993. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Lu, C.; Huang, Y.; Zhou, J.; Wang, X.; Liu, C.; Chen, J.; Le, H. SPINK1 promotes cell growth and metastasis of lung adenocarcinoma and acts as a novel prognostic biomarker. BMB Rep. 2018, 51, 648–653. [Google Scholar] [CrossRef] [Green Version]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [Green Version]
- Raponi, M.; Zhang, Y.; Yu, J.; Chen, G.; Lee, G.; Taylor, J.M.; Macdonald, J.; Thomas, D.; Moskaluk, C.; Wang, Y.; et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006, 66, 7466–7472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Leite de Oliveira, R.; Wang, C.; Fernandes Neto, J.M.; Mainardi, S.; Evers, B.; Lieftink, C.; Morris, B.; Jochems, F.; Willemsen, L.; et al. High-Throughput Functional Genetic and Compound Screens Identify Targets for Senescence Induction in Cancer. Cell Rep. 2017, 21, 773–783. [Google Scholar] [CrossRef] [Green Version]
- Lemoine, N.R.; Staddon, S.; Dickson, C.; Barnes, D.M.; Gullick, W.J. Absence of activating transmembrane mutations in the c-erbB-2 proto-oncogene in human breast cancer. Oncogene 1990, 5, 237–239. [Google Scholar]
- Lemoine, N.R.; Wyllie, F.S.; Lillehaug, J.R.; Staddon, S.L.; Hughes, C.M.; Aasland, R.; Shaw, J.; Varhaug, J.E.; Brown, C.L.; Gullick, W.J.; et al. Absence of abnormalities of the c-erbB-1 and c-erbB-2 proto-oncogenes in human thyroid neoplasia. Eur. J. Cancer 1990, 26, 777–779. [Google Scholar] [CrossRef]
- Soussi, T.; Wiman, K.G. TP53: An oncogene in disguise. Cell Death Differ. 2015, 22, 1239–1249. [Google Scholar] [CrossRef]
- Yip, S.C.; Saha, S.; Chernoff, J. PTP1B: A double agent in metabolism and oncogenesis. Trends Biochem. Sci. 2010, 35, 442–449. [Google Scholar] [CrossRef] [Green Version]
- Shen, L.; Shi, Q.; Wang, W. Double agents: Genes with both oncogenic and tumor-suppressor functions. Oncogenesis 2018, 7, 25. [Google Scholar] [CrossRef]
- Sahm, F.; Koelsche, C.; Meyer, J.; Pusch, S.; Lindenberg, K.; Mueller, W.; Herold-Mende, C.; von Deimling, A.; Hartmann, C. CIC and FUBP1 mutations in oligodendrogliomas, oligoastrocytomas and astrocytomas. Acta Neuropathol. 2012, 123, 853–860. [Google Scholar] [CrossRef]
- Debaize, L.; Troadec, M.B. The master regulator FUBP1: Its emerging role in normal cell function and malignant development. Cell Mol. Life Sci. 2019, 76, 259–281. [Google Scholar] [CrossRef]
- Zhao, Y.; Hu, X.; Liu, Y.; Dong, S.; Wen, Z.; He, W.; Zhang, S.; Huang, Q.; Shi, M. ROS signaling under metabolic stress: Cross-talk between AMPK and AKT pathway. Mol. Cancer 2017, 16, 79. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, C.; Sueyoshi, Y.; Ema, M.; Mori, Y.; Takaishi, K.; Hisatomi, H. Induction of oxidative stress by anticancer drugs in the presence and absence of cells. Oncol. Lett. 2017, 14, 6066–6070. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Chung, H.J.; Vogt, M.; Jin, Y.; Malide, D.; He, L.; Dundr, M.; Levens, D. JTV1 co-activates FBP to induce USP29 transcription and stabilize p53 in response to oxidative stress. EMBO J. 2011, 30, 846–858. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, M.; Kim, H.J.; Kim, T.-J.; Byun, J.-S.; Lee, J.-H.; Lee, D.H.; Kim, W.; Kim, D.-Y. Multiple Functions of Fubp1 in Cell Cycle Progression and Cell Survival. Cells 2020, 9, 1347. https://doi.org/10.3390/cells9061347
Kang M, Kim HJ, Kim T-J, Byun J-S, Lee J-H, Lee DH, Kim W, Kim D-Y. Multiple Functions of Fubp1 in Cell Cycle Progression and Cell Survival. Cells. 2020; 9(6):1347. https://doi.org/10.3390/cells9061347
Chicago/Turabian StyleKang, Mingyu, Hyeon Ji Kim, Tae-Jun Kim, Jin-Seok Byun, Jae-Ho Lee, Deok Heon Lee, Wanil Kim, and Do-Yeon Kim. 2020. "Multiple Functions of Fubp1 in Cell Cycle Progression and Cell Survival" Cells 9, no. 6: 1347. https://doi.org/10.3390/cells9061347