Abstract
Background
Anti-Müllerian hormone (Amh) plays a critical role in both early sex determination and later gonad development in vertebrate species. However, it remains unknown in northern snakehead (Channa argus), which is economically important freshwater fish with sexual dimorphism.
Objective
This study aimed to identify the expression profiles and estrogenic regulation of CaAmh during gonadal sex differentiation in C. argus.
Methods
The cDNA and genomic DNA sequences of CaAmh were identified by PCR and RACE techniques. The expression patterns of CaAmh were detected by qRT-PCR during the gonadal sex differentiation and after 17α-ethinyloestradiol (EE2) treatments.
Results
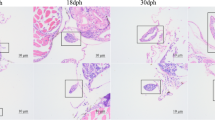
CaAmh is composed of seven exons and six introns, and its full-length cDNA is 2413 bp in length, with 1635 bp open reading frame (ORF) that encodes a 544 amino acid protein. Tissues expression patterns revealed that CaAmh display the highest expression in testis of XY males (40.36 folds, p < 0.01). The spatio-temporal expression patterns during gonadal sex differentiation indicated that CaAmh expression differed between XX females and XY males at 30 day after hatching (dah), and reached to the peak (36.03 folds, p < 0.01) at 90 dah in XY gonads. However, CaAmh expression in XX gonads remained low throughout the sampling period. Furthermore, CaAmh expression in the gonads (ovaries) of the sex-reversed XY fish (XY-F) by the administration of estrogen EE2 was downregulated to low level, similar to that in ovaries of normal XX females (XX-F).
Conclusions
These results show that Amh plays a critical role in testicular differentiation of C. argus and it is apparently modulated by estrogens in this species.




Similar content being viewed by others
References
Anttonen M, Unkilakallio L, Leminen A et al (2005) High GATA-4 expression associates with aggressive behavior, whereas low anti-Müllerian hormone expression associates with growth potential of ovarian granulosa cell tumors. J Clin Endocrinol Metab 90:6529–6535. https://doi.org/10.1210/jc.2005-0921
Behringer RR, Finegold MJ, Cate RL (1994) Müllerian-inhibiting substance function during mammalian sexual development. Cell 79:415–425. https://doi.org/10.1016/0092-8674(94)90251-8
Bej DK, Miyoshi K, Hattori RS et al (2017) A duplicated, truncated amh gene is involved in male sex determination in an old world silverside. G3 Genes Genomes Genet 7:2489–2495. https://doi.org/10.1534/g3.117.042697
Blasco M, Fernandino JI, Guilgur LG et al (2010) Molecular characterization of cyp11a1 and cyp11b1 and their gene expression profile in pejerrey (Odontesthes bonariensis) during early gonadal development. Comp Biochem Physiol A Mol Integr Physiol 156:110–118. https://doi.org/10.1016/j.cbpa.2010.01.006
Fernandino JI, Hattori RS, Kimura H et al (2008) Expression profile and estrogenic regulation of anti-müllerian hormone during gonadal development in pejerrey Odontesthes bonariensis, a teleost fish with strong temperature-dependent sex determination. Dev Dyn 237:3192–3199. https://doi.org/10.1002/dvdy.21731
Halm S, Rocha A, Miura T et al (2007) Anti-Müllerian hormone (AMH/AMH) in the European sea bass: Its gene structure, regulatory elements, and the expression of alternatively-spliced isoforms. Gene 388:148–158. https://doi.org/10.1016/j.gene.2006.10.018
Han Y, Zhao M, Wang L et al (2019) Overexpression of Anti-müllerian hormone gene in vivo affects Gonad sex differentiation in undifferentiated orange-spotted groupers (Epinephelus coioides). Front Endocrinol (Lausanne) 10. https://doi.org/10.3389/fendo.2019.00210
Hattori RS, Murai Y, Oura M et al (2012) A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proc Natl Acad Sci U S A 109:2955–2959. https://doi.org/10.1073/pnas.1018392109
He Y, Wang X, Wu X et al (2018) Expression profiles of amh and foxl2 in Schizothorax kozlovi, and their response to temperature during the early developmental stage. J Genet 97:127–136. https://doi.org/10.1007/s12041-018-0889-9
Johnsen H, Tveiten H, Torgersen J, Andersen O (2013) Divergent and sex-dimorphic expression of the paralogs of the Sox9-Amh-Cyp19a1 regulatory cascade in developing and adult atlantic cod (Gadus morhua L.). Mol Reprod Dev 80:358–370. https://doi.org/10.1002/mrd.22170
Kluver N, Pfennig F, Pala I et al (2007) Differential expression of anti-Müllerian hormone (amh) and anti‐Müllerian hormone receptor type II (amhrII) in the teleost medaka. Dev Dyn 236:271–281. https://doi.org/10.1002/dvdy.20997
Kwan TN, Patil JG (2019) Sex biased expression of anti-Mullerian hormone (amh) gene in a live bearing fish, Gambusia holbrooki: Evolutionary implications and potential role in sex differentiation. Comp Biochem Physiol Part B Biochem Mol Biol 231:59–66. https://doi.org/10.1016/j.cbpb.2019.02.004
Li M, Sun Y, Zhao J et al (2015) A tandem duplicate of anti-Müllerian hormone with a missense SNP on the Y chromosome is essential for male sex determination in Nile Tilapia, Oreochromis niloticus. PLOS Genet 11. https://doi.org/10.1371/journal.pgen.1005678
Lin Q, Mei J, Li Z et al (2017) Distinct and cooperative roles of amh and dmrt1 in self-renewal and differentiation of male germ cells in zebrafish. Genetics 207:1007–1022. https://doi.org/10.1534/genetics.117.300274
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Newhard JJ (2015) Identification and location of testes in the invasive Channa argus cantor (northern snakehead). Northeast Nat 22. https://doi.org/10.1656/045.022.0306
Ou M, Yang C, Luo Q et al (2017) An NGS-based approach for the identification of sex-specific markers in snakehead (Channa argus). Oncotarget 8:98733–98744. https://doi.org/10.18632/oncotarget.21924
Ou M, Zhao J, Luo Q et al (2018) Characteristics of hybrids derived from Channa argus ♀ × Channa maculata ♂. Aquaculture 492:349–356. https://doi.org/10.1016/J.AQUACULTURE.2018.04.038
Pfennig F, Standke A, Gutzeit HO (2015) The role of Amh signaling in teleost fish—multiple functions not restricted to the gonads. Gen Comp Endocrinol 223:87–107. https://doi.org/10.1016/j.ygcen.2015.09.025
Poonlaphdecha S, Pepey E, Huang S et al (2011) Elevated amh gene expression in the brain of male Tilapia (Oreochromis niloticus) during testis differentiation. Sex Dev 5:33–47. https://doi.org/10.1159/000322579
Rey R, Lukascroisier C, Lasala C, Bedecarras P (2003) AMH/MIS: what we know already about the gene, the protein and its regulation. Mol Cell Endocrinol 211:21–31. https://doi.org/10.1016/j.mce.2003.09.007
Rodriguezmari A, Yan Y, Bremiller R et al (2005) Characterization and expression pattern of zebrafish anti-Mullerian hormone (amh) relative to sox9a, sox9b, and cyp19a1a, during gonad development. Gene Expr Patterns 5:655–667. https://doi.org/10.1016/j.modgep.2005.02.008
Schepers G, Wilson MJ, Wilhelm D, Koopman P (2003) SOX8 is expressed during testis differentiation in mice and synergizes with SF1 to activate the Amh promoter in vitro. J Biol Chem 278:28101–28108. https://doi.org/10.1074/jbc.M304067200
Schulz RW, Bogerd J, Male R et al (2007) Estrogen-induced alterations in amh and dmrt1 expression signal for disruption in male sexual development in the zebrafish. Environ Sci Technol 41:6305–6310. https://doi.org/10.1021/es070785+
Takada S, Mano H, Koopman P (2005) Regulation of Amh during sex determination in chickens: Sox gene expression in male and female gonads. Cell Mol Life Sci 62:2140–2146. https://doi.org/10.1007/s00018-005-5270-5
Visser JA, Themmen APN (2005) Anti-Müllerian hormone and folliculogenesis. Mol Cell Endocrinol 234:81–86. https://doi.org/10.1016/j.mce.2004.09.008
Wang D-D, Zhang G-R, Wei K-J et al (2015) Molecular identification and expression of the Foxl2 gene during gonadal sex differentiation in northern snakehead Channa argus. Fish Physiol Biochem 41:1419–1433. https://doi.org/10.1007/s10695-015-0096-z
Western PS, Harry JL, Graves JAM, Sinclair AH (1999) Temperature-dependent sex determination in the American alligator: AMH precedes SOX9 expression. Dev Dyn 216:411–419. https://doi.org/10.1002/(SICI)1097-0177(199912)216:4/5<411::AID-DVDY9>3.0.CO;2-Y
Wu G, Li H, Tey W et al (2017) Expression profile of amh/Amh during bi-directional sex change in the protogynous orange-spotted grouper Epinephelus coioides. PLoS One 12. https://doi.org/10.1371/journal.pone.0185864
Xu J, Bian C, Chen K et al (2017) Draft genome of the northern snakehead, Channa argus. Gigascience 6:1–5. https://doi.org/10.1093/gigascience/gix011
Yamamoto Y, Zhang Y, Sarida M et al (2014) Coexistence of genotypic and temperature-dependent sex determination in Pejerrey Odontesthes bonariensis. PLoS One 9. https://doi.org/10.1371/journal.pone.0102574
Yoshinaga N, Shiraishi E, Yamamoto T et al (2004) Sexually dimorphic expression of a teleost homologue of Müllerian inhibiting substance during gonadal sex differentiation in Japanese flounder, Paralichthys olivaceus. Biochem Biophys Res Commun 322:508–513. https://doi.org/10.1016/j.bbrc.2004.07.162
Yue Z, Gao S, Deng F et al (1996) A histological study on the ovary development of Channa argus. J Wuhan Univ Sci Ed 42:225–232. https://doi.org/10.14188/j.1671-8836.1996.02.018
Acknowledgements
This work was supported by the Central Public-interest Scientific Institution Basal Research Fund, CAFS (Grant number 2017HY-ZC0404), China Agriculture Research System (Grant number CARS-46), the National Natural Science Foundation of China (Grant number 31902351), the Central Public-interest Scientific Institution Basal Research Fund, CAFS (Grant number 2020TD34), the Pearl River S&T Nova Program of Guangzhou (no. 201710010174) and National Key R&D Program of China (Grant number 2018YFD0901201).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
13258_2020_943_MOESM1_ESM.pdf
Supplementary file1 Fig. S1. Nucleotide and putative amino sequences of CaAmh. The sequences numbers of nucleotide (lower row) and putative amino acid (upper row) are shown on the left. The translation initiation codon (ATG), stop codons (TAA) are in bold and red. The motif associated mRNA instability (ATTTA) is shown as double underscore, and poly-adenylation signal sequence (AATAA) is shown as wavy line. The signal peptide is showd as underscore. The AMH-N terminal is marked with green background, and TGF-β domain is highlighted in green background. Fig. S2. Multiple alignments of CaAmh with Amh proteins from various species. The amino acid sequences of Amhs from typical organisms were aligned using the ClustalW 2.1 program. The black shade represent 100% identity, dark gray represented 80% identity, and TGF-β domain is marked in red box. Fig. S3. Phylogenetic relationship between the Amh proteins in different species. A neighbor-joining phylogenetic tree was constructed using MEGA 5.0 software. The bootstrap values of the branches were obtained by testing the tree 1000 times and values were over 50% percent marked. The GenBank accession numbers of Amh proteins are given in Table 2 (PDF 474 kb)
Rights and permissions
About this article
Cite this article
Luo, Q., Ou, M., Zhao, J. et al. Expression profile and estrogenic regulation of Amh during gonadal sex differentiation in northern snakehead (Channa argus). Genes Genom 42, 827–835 (2020). https://doi.org/10.1007/s13258-020-00943-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-020-00943-7