Abstract
Background
Bacterial leaf streak (BLS) and bacterial blight (BB) are two major prevalent and devastating rice bacterial diseases caused by the Gram-negative bacteria of Xanthomonas oryzae pv. oryzicola (Xoc) and Xanthomonas oryzae pv. oryzae (Xoo), respectively. Previously, we identified a defence-related (DR) gene encoding a small heat shock protein, OsHsp18.0-CI, that positively regulates BLS and BB resistance in rice.
Results
To reveal the regulatory mechanism of the OsHsp18.0-CI response to Xoc and Xoo, we characterized the class B heat shock factor (Hsf), OsHsfB4d, through transcriptional analysis and a transgenic study. OsHsfB4d is upregulated post inoculation by either the Xoc strain RS105 or Xoo strain PXO99a in Zhonghua 11 (wild type, ZH11) as well as in OsHsp18.0-CI overexpressing rice plants. Transient expression of OsHsfB4d can activate the expression of green fluorescent protein (GFP) and luciferase (Luc) via the OsHsp18.0-CI promoter. Rice plants overexpressing OsHsfB4d exhibited enhanced resistance to RS105 and PXO99a as well as increased expression of OsHsp18.0-CI and pathogenesis-related genes. Furthermore, we found that OsHsfB4d directly binds to a DNA fragment carrying the only perfect heat shock element (HSE) in the promoter of OsHsp18.0-CI.
Conclusion
Overall, we reveal that OsHsfB4d, a class B Hsf, acts as a positive regulator of OsHsp18.0-CI to mediate BLS and BB resistance in rice.
Similar content being viewed by others
Background
Rice is an important staple crop worldwide that represents 40% of total grain output and nearly 60% of global food consumption (Sharma et al. 2012). However, it has been shown to suffer more than 70 diseases caused by fungi, bacteria, viruses and nematodes during rice growth (Niño-Liu et al. 2006; Ke et al. 2017). There are two major bacterial diseases, bacterial blight (BB) and bacterial leaf streak (BLS), caused by the gram-negative bacteria Xanthomonas oryzae that frequently occur in rice. BB is caused by Xanthomonas oryzae pv. oryzae, which enters into the rice leaf through hydathodes or wounds and colonizes in the xylem vessels. But BLS is caused by Xanthomonas oryzae pv. oryzicola (Xoc), which penetrates into the leaf through stomata or wounds and colonizes the intercellular space of leaf tissue and finally results in water-soaked stripe lesions (Niño-Liu et al. 2006; Ke et al. 2017; Ju et al. 2017). Currently, BB is well studied for host resistance. Over 40 major resistance genes and 30 defense-related (DR) genes were identified to control the race-specific or spectrum resistance to Xoo isolates (Ke et al. 2017; Ju et al. 2017). BLS is becoming a major concern due to its high prevalence and seriously affecting the yield and quality of rice production (Xu et al. 2010; Zhang et al. 2015). To date, only the Xo1 locus, which encodes a putative receptor, has been reported to confer qualitative resistance against the African clade of Xoc strains but not Asia strains (Triplett et al. 2016). Otherwise, BLS resistance was considered to be controlled by a number of quantitative trait loci (QTLs), such as qBlsr5a, which may consist of xa5, OsPGIP1 and OsPGIP4 from the rice variety Acc8558 (Xie et al. 2014; Feng et al. 2016; Wu et al. 2019).
Several DR genes have been demonstrated to exhibit upregulated expression upon Xoc inoculation and to positively or negatively regulate the rice BLS resistance (Xu et al. 2013). Overexpression of the mitogen-activated protein kinase gene OsMPK6 increased susceptibility to Xoc strain RS105, implying the negative regulation of rice BLS resistance (Shen et al. 2010). Consistent with the negative regulation of rice immunity, suppression of DR genes, such as OsWRKY45–1 (Tao et al. 2009), OsDEPG1 (Guo et al. 2012), OsNRRB (Guo et al. 2014), OsImpα1a and OsImpα1b (Hui et al. 2019), could enhance the resistance to Xoc. In contrast, other DR genes, including OsGH3–2 (Fu et al. 2011), OsPGIP4 (Feng et al. 2016), RGA4/RGA5 (Hutin et al. 2016), OsHsp18.0-CI (Ju et al. 2017), XCRK (Zhang et al. 2017), MPKK10.2 (Ma et al. 2017), OsPSKR1 (Yang et al. 2019) and OsPGIP1 (Wu et al. 2019), may positively regulate BLS resistance in rice. However, constitutively expressed of DR genes often result in enhanced resistance as well as impairment of agronomic traits such as yield and quality (Wiesner-Hanks et al. 2018). Therefore, understanding the transcriptional regulation mechanisms of DR genes and fine-tuning their regulation ability is critical and ideal for breeding resistant rice varieties.
Heat shock proteins (Hsps) are conserved across a wide diversity of organisms. They are chaperones that assist in protein folding and prevent irreversible protein aggregation (Waters 2012). Commonly, the transcription of a heat shock protein gene is regulated by heat shock factors (Hsfs) in plants. Plants contain multimember Hsf gene families organized into evolutionarily conserved and structurally distinct A, B and C classes (Kotak et al. 2004; Scharf et al. 2012). The modular Hsf structure comprises an N-terminal DNA binding domain (DBD), an adjacent oligomerization domain (OD or HR-A/B region), nuclear localization and export signals (NLS/NES) and a C-terminal activation domain (CTAD) (Lavania et al. 2018). Class A Hsfs specifically contain a unique C-terminal activation domain with aromatic hydrophobic acidic (AHA) motifs, while class B and class C Hsfs are characterized by the lack of an activation domain (Nover et al. 2001; Baniwal et al. 2004). Normally, Hsfs activates or inhibits the expression of the targeted Hsp gene by directly binding to the heat shock element (HSE), a consensus sequence of 5′-nGAAn-3′ embedded in the promoter (Sakurai and Enoki 2010; Scharf et al. 2012). Class A Hsfs are generally considered positive regulators, while class B and C Hsfs are considered negative regulators.
Currently, the role of Hsfs in basal tolerance to heat and other abiotic stresses is well established in plants (Hahn et al. 2011; Scharf et al. 2012). However, the role of Hsfs in plant disease resistance is poorly understood. Overexpression of AtHsfA1b resulted in increased resistance to Pseudomonas syringae (Pst) DC3000 and Hyaloperonospora Arabidopsis pv. WACO9 (Hpa) in Arabidopsis (Bechtold et al. 2013). SlHsfA1a positively regulated Wfi1, an RBOH gene that triggers the production of reactive oxygen species (ROS) in the apoplast during Meloidogyne incognita infection of the roots of tomato plants (Zhou et al. 2018). In contrast, class B Hsf members play a negative regulatory role in disease resistance mechanisms. AtHsfB1 and AtHsfB2b negatively regulate the expression of the plant defensin genes PDF1.2a and b (Kumar et al. 2009). As a result, the single mutant AtHsfB2b and the double mutant AtHsfB1/AtHsfB2b exhibited resistance to the necrotrophic fungus Alternaria brassicicola (Liu and Charng 2013). In addition, overexpressing the grape VpHsf1 gene (encoding a member of the B family) in tomatoes can increase the sensitivity of plants to Erysiphe necator (Peng et al. 2013).
Rice contains a total of 25 typical Hsfs members in its genome. Among them, 22, 10 and 14 OsHsfs have been identified that respond to heat, chilling and oxidative stress, respectively (Mittal et al. 2009). However, the functions of most OsHsfs have not been revealed. Previously, we identified a small heat shock protein encoded by OsHsp18.0-CI that positively regulates rice BLS and BB resistance (Ju et al. 2017; Zuo et al. 2019). Using an RNA-seq strategy, a number of differentially expressed genes (DEGs) involved in basal defence were shown to be enriched in OsHsp18.0-CI-overexpressing (OE) lines compared with wild-type ZH11 (Ju et al. 2017; Zuo et al. 2019). Among the characterized DEGs, one of the candidate Hsfs, OsHsfB4d, was identified by deeply analysing the comparative transcriptome data and investigating the regulatory interaction with OsHsp18.0-CI. Importantly, we found that OsHsfB4d acts as a positive regulator to activate the expression of OsHsp18.0-CI by directly targeting its promoter sequence.
Results
The Expression of OsHsfB4d is Induced by Xanthomonas Oryzae pv. Oryzae and X. Oryzae pv. Oryzicola
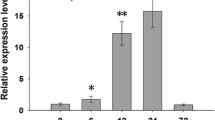
As Hsfs specifically regulate the expression of Hsp genes, to reveal the regulatory mechanism of OsHsp18.0-CI in rice immunity, we analysed the transcriptome data from rice lines overexpressing OsHsp18.0-CI cDNA under the control of the Ubi promoter post Xoc or Xoo inoculation (Ju et al. 2017; Zuo et al. 2019). As shown in the supporting file Figure S1, we investigated the transcriptional data for all class A and B Hsfs. Several Hsfs were upregulated in transgenic plants or by pathogen inoculation, including OsHsfA2f, OsHsfA7, OsHsfB1 and OsHsfB4d. OsHsfB4d attracted attention because its expression was significantly induced by both the Xoc and Xoo strains. The transcription level of OsHsfB4d was approximately 8-fold increased compared with that in the control 24 h post inoculation (hpi) with Xoc strain RS105. OsHsfB4d was also significantly upregulated in OsHsp18.0-CI OE plants compared with wild-type ZH11 plants without inoculation with RS105 (Fig. 1a). Upregulation pattern for OsHsfB4d was also identified in response to the Xoo strain PXO99 (Fig. 1a). The results of the RNA-seq data analysis were further validated by qRT-PCR to confirm the induction of OsHsfB4d expression during Xoc and Xoo inoculation (Fig. 1b, c). The above results demonstrated that increased level of transcripts of OsHsfB4d was identified in ZH11 rice after the two bacterial pathogens challenge, suggesting that OsHsfB4d may be involved in the resistance of rice to these two pathogens.
OsHsfB4d expression is induced by Xanthomonas oryzae pv. oryzicola (Xoc) and Xanthomonas oryzae pv. oryzae (Xoo) inoculation. a Hierarchical cluster analysis of OsHsfB4d using the OsHsp18.0-CI RNA-seq data. Quantification of the expression level of OsHsfB4d in response to RS105 (b) or PXO99a (c) by qRT-PCR. Fully expanded leaves of six-week-old ZH11 and OsHsp18.0-CI-OE plants were harvested at 0 and 24 h post inoculation. The bars represent the mean (three replicates for gene expression) ± SD. The significance of expression compared to that at 0 h in the WT at P values of less than 0.05 and 0.01 are indicated by “*” and “**”, respectively
OsHsfB4d Transiently Activated the Expression of OsHsp18.0-CI
To evaluate whether OsHsfB4d could regulate the expression of OsHsp18.0-CI, a transient expression assay was performed by coexpressing OsHsfB4d and green fluorescent protein (GFP) or luciferase (LUC) by using OsHsp18.0-CI promoter. As shown in Fig. 2a, different combinations of constructs were transiently introduced into Nicotiana benthamiana leaves by Agrobacterium tumefaciens-mediated transformation, and GFP activity was assayed 24 h after infiltration. No obvious fluorescence was detected in the control plant leaves, whereas significant GFP fluorescence was observed in OsHsfB4d plants with reporter constructs (Fig. 2b). The quantification of the GFP fluorescence intensity was performed to obtain a similar result (Fig. 2c). In addition, coexpression with OsHsfB4d also activated the fluorescence of the OsHsp18.0-CIpro:LUC construct (Fig. 2d). These results indicated that OsHsfB4d might act as a regulator and activate the expression of OsHsp18.0-CI in planta.
Fluorometric assays of GFP or LUC driven by OsHsp18.0-CI promoter constructs. a Diagram of the constructs of OsHsfB4d and the OsHsp18.0-CI promoter. b GFP fluorescence of the OsHsp18.0-CI promoter DNA construct (OsHsp18.0-CIPro:GFP). Coexpression with or without the OsHsfB4d in N. benthamiana. Images were photographed at 2 d after infiltration. Bar = 200 μm. c Quantitative analysis of the fluorescence intensity. The results are presented as the average fluorescence intensity using ZEN software. The experiments were repeated three times. ** indicates significant differences according to Fisher’s least-significant difference test (P < 0.01). d Luciferase assay of the co-expressed OsHsfB4d and the promoters of OsHsp18.0-CI (OsHsp18.0-CIPro:LUC). Mock represents the 10 mM MgCl2 solution
OsHsfB4d-Overexpressing Lines Conferred Enhanced Disease Resistance to BLS and BB
To investigate the role of OsHsfB4d in the disease resistance response, an overexpression vector expressing OsHsfB4d (OE) was constructed and introduced into the rice variety ZH11 (Fig. 3a). From the T2 generation, three homozygous transgenic lines, OE3, OE10 and OE13, were selected for further investigation. The transcription level of OsHsfB4d was increased approximately 249.2-fold, 82.8-fold and 172.5-fold in OE3, OE10 and OE13 compared to that in the WT, respectively (Fig. 3b). The increased content of OsHsfB4d protein was also confirmed by a western blotting assay with a c-Myc-tagged antibody (Fig. 3c). The fully expanded leaves of six-week-old plants were used for disease assessment by inoculation with RS105. The lesion length in the WT was recorded as being 1.98 ± 0.21 cm on average. Compared to the WT, the three OE lines showed shorter average lesion lengths of 1.19 ± 0.22 cm, 0.94 ± 0.19 cm and 1.15 ± 0.27 cm for OE3, OE10 and OE13, respectively (Fig. 3d, e). Consistently, though there are no significant difference between WT and OE lines at 7 dpi, the bacterial populations in the leaves of the OE lines were significantly decreased compared to those in the WT at 14 dpi (Fig. 3f). Overall, these results suggested that overexpression of OsHsfB4d enhanced rice resistance to RS105.
Resistance performance of transgenic OsHsfB4d OE plants from the T2 generation. a Diagram of the OsHsfB4d overexpression construct. b Relative expression levels of OsHsfB4d in three identified T2 transgenic lines. OsActin was used as an internal control. Bars represent the mean (three replicates for gene expression) ± SD. c Immunoblot analysis of OsHsfB4d in transgenic lines. Total protein extracts were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and probed with antibodies against the c-Myc tag (α-Myc). The stained Rubisco protein shows that equal amounts of protein sample were loaded. The immunoblot band shows OsHsfB4d-Myc of approximately 35 kD. d Representative lesion sites in OsHsfB4d OE leaves at 14 dpi with Xoc strain RS105. e Lesion lengths scored for T2 transgenic plants with pCXUN::OsHsfB4d. Eight individuals were scored for each line and the WT. “**” indicates significant (t test, P < 0.01) differences (n = 16). f Bacterial population growth in leaves from OsHsfB4d OE and WT plants inoculated with RS105. Colonies were counted with the leaf segment up to 5 cm from the inoculation site. Error bars represent the standard deviation of three independent leaves. The bars represent the mean ± SD. “**” indicates significant (t test, P < 0.01) differences. g The phenotypes of the PXO99a lesions that developed on ZH11 and OsHsfB4d-OE lines at 14 dpi. h Lesion lengths were scored for OsHsfB4d-OE lines (n = 10). Asterisks indicate significant difference between transgenic and wildtype (WT) plants at **P < 0.01 or *P < 0.05
We also investigated the BB resistance for the OE lines. As shown in Fig. 3g, the lesion lengths of all three OE lines for PXO99a at 14 dpi were measured as 8.83 ± 3.05 cm, 7.75 ± 1.81 cm and 6.25 ± 0.50 cm, respectively (Fig. 3h). Which showed more shorter lesion lengths compared with WT (12.00 ± 3.55 cm). Therefore, OsHsfB4d could be considered as a positive regulator involved in the response to Xoc and Xoo.
Activated Expression of Pathogenesis-Related (PR) Genes in OsHsfB4d OE Lines
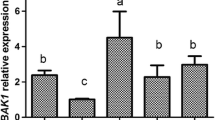
As a candidate regulator of OsHsp18.0-CI, we first investigated the transcription level of OsHsp18.0-CI in rice lines OE3, OE10 and OE13. As shown in Fig. 4a, the relative expression of OsHsp18.0-CI was 10.6-, 13.1-, and 5.9- fold higher in OE3, OE10 and OE13 than in WT, respectively. Previous studies demonstrated that pathogenesis-related (PR) genes are induced in OsHsp18.0-CI overexpression lines without inoculation with RS105 (Ju et al. 2017). Then, three pathogenesis-related genes were measured with qRT-PCR to determine the basal levels of their transcripts in the OsHsfB4d-OE lines. All PR genes were increased expression in three OE lines compared with the WT. As shown in Fig. 4b-d, both OsPR1a and OsPR1b were significantly upregulated, and OsPAL1 was slightly upregulated in one of the OE lines (OE10) compared to the WT. In addition, the increased levels of PR genes coincided with the increased level of OsHsp18.0-CI expression and the resistance performance of the three OE lines. These results indicated that the OE lines may enhance BLS resistance by activating the expression of OsHsp18.0-CI and PR genes.
Overexpressing OsHsfB4d activates the expression of OsHsp18.0-CI and pathogenesis-related genes. Quantitative RT-PCR was performed for OsHsp18.0-CI(a) and three PR genes, including OsPR1a(b), OsPR1b(c) and OsPAL1(d), in 6-week-old wild-type and OsHsfB4d overexpression lines. The data are shown as the mean ± SD of three biological replicates. The experiments were repeated two times with similar results
Knockout of OsHsfB4d has no Effect on Resistance to BLS and BB
To further identify the role of OsHsfB4d in rice-Xoc interactions, we generated CRISPR-Cas9 OsHsfB4d knockout lines in ZH11. Three independent individuals, CAS10, CAS16 and CAS22, were identified to have different null mutations and displayed various indel polymorphisms (Fig. 5a, Fig. S2). The CAS lines were assessed the resistance to RS105 in the T2 generations. At 14 dpi, the three knockout lines exhibited lesion lengths of 1.89 ± 0.16 cm, 1.95 ± 0.21 cm and 2.11 ± 0.21 cm, respectively, which were similar to the average of 1.98 ± 0.21 cm for WT plants (Fig. 5b, c). Consistent with these results, the bacterial populations in leaves were essentially unchanged between the CAS lines and WT at 7 dpi or 14 dpi (Fig. 5d). Additionally, the expression of OsHsp18.0-CI in the CAS lines were not significantly different from that in the WT (Fig. 5e).
Resistance performance of transgenic OsHsfB4d-cas9 plants from the T2 generation. a Diagram of the Cas9 enzyme cleavage site and the genome editing of OsHsfB4d. Three mutation types, including a 1 base deletion, a 1 base insertion and a 4 bases deletion, were detected in the three transgenic lines. b Representative lesion sites from OsHsfB4d-cas9 plants at 14 dpi with Xoc strain RS105. c Lesion lengths in the OsHsfB4d-cas9 lines. Each line comprises seven individual transgenic plants assessed in this experiment at 6-week-old stage. “n.s.” means not significant according to the statistical analysis (n = 10). d Bacterial population growth in leaves of OsHsfB4d-cas9 and WT plants inoculated with RS105. The bars represent the mean ± SD. “n.s.”: not significant. e The transcript detection of OsHsp18.0-CI in 6-week-old wild-type and OsHsfB4d-cas9 lines. The bars represent the mean ± SD. f The phenotypes of the PXO99a lesions that developed on ZH11 and OsHsfB4d-cas9 lines at 14 dpi. g Lesion lengths were scored for OsHsfB4d-cas9 lines (n = 10). “n.s.”: not significant
By the BB inoculation, the lesion lengths between three knockout lines (CAS10, CAS16 and CAS22) and WT also did not changed significantly (Fig. 5f), the statistical lesion lengths were 11.4 ± 4.03 cm, 11.73 ± 4.08 cm, 10.43 ± 3.32 cm, 12 ± 3.55 cm, respectively (Fig. 5g). Overall, we concluded that the deletion of OsHsfB4d had little effect on the rice-Xanthomonas interaction.
OsHsfB4d Directly Targets the Promoter of OsHsp18.0-CI
Hsfs usually regulate the expression of Hsp genes by directly targeting the HSE element to mediate physiological processes (Scharf et al. 2012). Nine putative HSEs, including one perfect HSE and eight imperfect HSEs, were previously predicted to be located in a 567 bp DNA fragment of the promoter of OsHsp18.0-CI by sequence analysis (Guan et al. 2004). To investigate the interaction between OsHsfB4d and the OsHsp18.0-CI promoter, a His-tagged recombinant OsHsfB4d protein was purified via prokaryotic expression and subsequently used to assess binding ability with electrophoretic mobility shift assays (EMSA). The results showed that OsHsfB4d could bind to DNA probe 1, which contained the perfect HSE motif but failed to bind DNA probe 2 and probe 3, which are located at other positions in OsHsp18.0-CI (Fig. 6a, b, Fig. S3). To further confirm the binding ability, we performed ChIP-qPCR assays using OE10 rice plants carrying OsHsfB4d-Myc. Consistent with the EMSA results, relatively greater enrichment of the probe 1 DNA fragment, containing the perfect HSE motif, was observed during immunoprecipitation than that of the DNA fragment of probe 2 or β-actin by using anti-Myc agarose beads (Fig. 6c). Based on the results of EMSA and ChIP-qPCR, we concluded that OsHsfB4d directly binds to the OsHsp18.0-CI promoter.
OsHsfB4d directly binds to the promoter of OsHsp18.0-CI. a Diagram of the positions of the primers and probes used in the EMSA and ChIP-qPCR experiments. b EMSA using recombinant His-tagged OsHsfB4d and the various deletion derivatives of the OsHsp18.0-CI promoter fragment. Lane 1 contained only the free probe. Lanes 2–7 contained the purified His-tagged OsHsfB4d and the probe. For the competitive EMSA, the purified OsHsfB4d protein was preincubated with 5×, 10×, 50×, 200× molar excess of unlabeled probe 1 (lane 3–6), 5× probe 2 (lane 8) and 5× probe 3 fragments (lane 10) before the addition of the corresponding biotin-labelled promoter fragment. c Binding of OsHsfB4d to the perfect HSE cis-element-containing fragment in a ChIP-qPCR assay. ChIP and EMSA experiment used the same primer pairs. The Actin was used as a nonspecific target gene
Considering the probe 1 DNA fragment bound to OsHsfB4d contains a perfect HSE motif and two imperfect ones, we further tested the interaction with shorter fragment containing the only perfect HSE motif. Firstly we fixed amount of labeled probes and uploaded different amount of proteins to detect the effect of the protein concentration on the protein-DNA interaction. The result showed the increase of bound probes with the increased amount of proteins (Fig. 7a), further confirmed the binding of probe 1 DNA fragment to OsHsfB4d. EMSA image demonstrated that OsHsfB4d protein bound the DNA probe containing the perfect HSE motif but failed to bind the similar DNA probe in which the HSE motif was mutated (Fig. 7b), indicating that OsHsfB4d specifically binds the perfect HSE motif of OsHsp18.0-CI promoter.
OsHsfB4d binds to the perfect HSE motif with the OsHsp18.0-CI promoter. a EMSA using the probe 1 and different amount of recombinant His-tagged OsHsfB4d. 1× represents about 0.53 μg of the purified OsHsfB4d. b OsHsfB4d bind to the perfect HSE cis-element. The OsHsfB4d protein was incubated with the labeled probe to serve as a positive control; mutated probes (Mu) were used as a negative control. 5-, 10-, 50- and 200-fold excesses of unlabeled probes were used for competition. 50- and 200-fold excesses of mutated probes were used for competition. Mutated probe in which the perfect HSE motif 5′- GAAACTTC − 3′ was replaced with 5′- CAAACAAT -3′
Discussion
Class B Hsfs are generally regarded as negative regulators that respond to abiotic stresses in plants. AtHsfB1 and AtHsfB2b have been shown to repress the induction of Hsps during stress recovery in Arabidopsis (Ikeda et al. 2011). Soybean HsfB2b inhibited GmNAC2 expression by directly binding to the HSE element in the salt stress response (Bian et al. 2020). To date, there have been few reports on the involvement of class B Hsfs in regulating the plant response to biotic stress. Furthermore, AtHsfB1 and AtHsfB2b have been shown to negatively regulate resistance to A. brassicicola by suppressing the expression of the plant defensin genes PDF1.2a and PDF1.2b (Kumar et al. 2009; Liu and Charng 2013). Do class B Hsfs act as positive regulators to directly activate the expression of targeted genes? In tomato, SlHsfB1 alone represses the transcription of Hsps, whereas it can positively regulate the transcription of Hsps by forming heterodimers with SlHsfA1 (Hahn et al. 2011). In addition to inhibiting GmNAC2 expression, HSFB2b also plays as a positive regulator that can bind to the promoter and activate the expression of GmC4H to promote flavonoid biosynthesis in response to salt stress in soybean (Bian et al. 2020). Here we also revealed that one of the class B Hsfs, OsHsfB4d, activates the expression of the target gene OsHsp18.0-CI (Fig. 4a) and enhances BLS and BB resistance (Fig. 3b-h) after overexpression in rice. Thus, we revealed a new scenario in which class B Hsfs may also act as positive regulators in response to biotic stresses.
In contrast to class A Hsfs, class B and C Hsfs lack the activation domain, so they are not complete transcription factors (Czarnecka-Verner et al. 2000, 2004; Kotak et al. 2004; Lavania et al. 2018). Previous studies demonstrated that the Hsfs of subclass A act as master regulators of the heat stress response (Mishra et al. 2002; Liu et al. 2011; Yoshida et al. 2011). Class B Hsfs usually play roles as co-activators or co-repressors (Czarnecka-Verner et al. 2000, 2004; Bharti et al. 2004; Ikeda et al. 2011; Schmidt et al. 2012). In this study, although we found that OsHsfB4d could directly bind to the promoter as well as activate the expression of OsHsp18.0-CI or GFP in OsHsfB4d OE lines (Fig. 4a) or in a co-transient expression system (Fig. 2b-d), it is still unclear whether OsHsfB4d could independently activate the expression of OsHsp18.0-CI. Because OsHsfB4d also lacks an activation domain and transactivation activity in yeast (Mittal et al. 2011; Lavania et al. 2018), we suggest that OsHsfB4d may act as a necessary cofactor in the transcriptional regulation of OsHsp18.0-CI, and the key regulator is conserved and exists in both rice and N. benthamiana.
There are 25 typical Hsfs genes in the rice genome. Of these, 13, 8 and 4 Hsfs belong to class A, class B and class C, respectively (Chauhan et al. 2011). We analysed all the transcription data for class A and class B Hsfs in the OsHsp18.0-CI OE lines post-inoculation with Xoc or Xoo. Hsfs exhibited various expression patterns in transgenic plants or after inoculation with different pathogens (Fig. S1), implying that they play different roles in the immune responses of rice. In this study, overexpression of OsHsfB4d enhanced resistance to the Xoc and Xoo (Fig. 3b-h), suggesting that OsHsfB4d positively mediates immunity in rice. Surprisingly, the three OsHsfB4d knockout lines exhibited no difference in BLS and BB resistance (Fig. 5b-g), implying that there are redundant activators that regulate the expression of OsHsp18.0-CI. Interestingly, the 567 bp-long promoter of OsHsp18.0-CI contains nine putative cis-elements of HSE, the only one perfect HSE element was confirmed to precisely bind to the OsHsp18.0-CI promoter (Fig. 7b) It would be available to be bound via additional HSE sites by other OsHsfs that need to cooperate with OsHsfB4d. Furthermore, this is a bidirectional promoter that drives the expression of OsHsp17.3 in an alternative orientation. The expression of OsHsp17.3 was induced in response to L-azetidine-2-carboxylic acid (AZC) and heat stress in rice and was coexpressed with OsHsfA4b in N.benthamiana (Guan et al. 2010). Therefore, the specific redundant activator of OsHsfB4d should be further investigated.
OsHsp18.0-CI has been identified to respond to treatment with anoxia, heat, cold, salt, drought, AZC, cadmium, and the bacterial pathogens Xoo and Xoc (Guan et al. 2004; Sarkar et al. 2009; Cui et al. 2013; Ham et al. 2013; Ju et al. 2017; Zuo et al. 2019). Several studies have indicated that overexpression of OsHsp18.0-CI enhances tolerance to abiotic and biotic stresses (Ham et al. 2013; Ju et al. 2017; Zuo et al. 2019). As OsHsfB4d is the upstream regulator of OsHsp18.0-CI, we showed that OsHsfB4d overexpression in rice enhanced BLS and BB resistance. It would be very interesting to expand the assessment to include more abiotic and biotic stressors in the future. OsHsfB4d maybe a good choice for breeding better rice varieties that are adapted to complex stress conditions in changed environments.
In conclusion, we showed that OsHsfB4d is a positive regulator involved in the rice-Xoc and rice-Xoo interaction. It could directly bind to the promoter and regulate the expression of OsHSP18.0-CI to enhance BLS and BB resistance in rice.
Methods
Plant Cultivation and Pathogen Inoculation
Seeds of the rice variety ZH11 and the transgenic lines were germinated and grown in a greenhouse at 28 ± 2 °C with 80% relative humidity and a 12-h photoperiod. The N.benthamiana plants used in this experiment were cultured in a chamber at 25 °C with a 16 h light / 8 h dark cycle. For inoculation, the Xoc strain RS105 and Xoo strain PXO99a was grown on PSA medium at 28 °C for 2 d and then suspended in 10 mM sterile MgCl2 at an OD600 = 0.5. For Xoc inoculation, at least five fully expanded leaves of 6-week-old plants were infiltrated at three positions by inoculation with a needleless syringe (Ju et al. 2017). The lengths of the lesions on the transgenic and wild type plants were scored at 14 dpi. For Xoo inoculation, 6-week-old rice plants were inoculated with PXO99a by the leaf-clipping method (Yang et al. 2016) and lesion lengths were measured at 14 dpi.
Vector Construction and Rice Transformation
The cDNAs of the LOC_Os03g25120 (OsHsfB4d) genes were amplified by RT-PCR using specific primers (Table S1). The OsHsfB4d fragment was cloned into the pCXUN-Myc vector under the control of the ubiquitin promoter (Chen et al. 2009). The CRISPR-OsHsfB4d vectors were generated according to the instructions (Ma et al. 2015). In brief, two sites for guide RNA (gRNA) targeted to the exon of OsHsfB4d were designed and transcribed from the U3 and U6a promoters, respectively, and subsequently cloned into the pYLCRISPR/Cas9-MH binary vectors. Then, the constructs were transferred into the Agrobacterium tumefaciens strain EHA105. All the constructs mentioned above were introduced into ZH11 using the standard Agrobacterium-mediated transformation system described previously (Li et al. 2013).
RNA Extraction and Real-Time RT-PCR
Pathogen-infected and control plant leaves were harvested at 24 h posttreatment for RNA isolation by using a Plant RNA Extraction Kit (Omega Bio-Tek, USA). First-strand cDNA was generated using the ReverTra Ace qPCR RT Master Mix with gDNA Remover kit (TOYOBO, Japan). Quantitative real-time PCR was performed on a qTOWER3G touch (Analytikjena, Germany) with KOD SYBR qPCR Mix (TOYOBO). The PCR program was performed as described in a previous study (Yang et al. 2019). The gene expression levels relative to those of the rice OsActin (LOC_Os03g50890) gene were analysed using the 2-△△Ct analysis method. The OsActin gene was used as an internal control to standardize the results. Gene expression levels were analysed by qRT-PCR assays, which were repeated at least twice in triplicate.
Fluorescence Microscopy Assay
Green fluorescent protein (GFP) was used as a reporter to investigate the expression of target gene fragments in planta. The pCXGFP-OsHsfB4d constructs were introduced into A. tumefaciens strain GV3101 and transiently expressed in N. benthamiana epidermis cells. GFP fluorescence was observed under a Leica M205 C stereomicroscope as described previously (Li et al. 2017; Yang et al. 2017). The fluorescence was quantified using an EnSpire Multimode Plate Reader (PerkinElmer, USA) as described previously (Yang et al. 2017).
Luciferase Reporter Assay
The constructs were transformed into Agrobacterium strain GV3101. Overnight cultures were collected by centrifugation, resuspended in MES buffer (10 mM MES pH 5.6, 10 mM MgCl2, and 150 mM acetosyringone) at an OD600 of 0.5 and incubated at room temperature for 2–3 h. The suspension was infiltrated into healthy leaves of 3-week-old N. benthamiana plants with a 2 mL needleless syringe. The plants were left under normal condition in chamber for 2 d after infiltration. Luciferin (1 mM) was infiltrated before the LUC signal was photographed with Night Shade LB985 (Berthold Technologies, Germany). The primer sequences used in this study are listed in Table S1.
EMSA
Full-length CDSs of OsHsfB4d were amplified and cloned into pET-28a.The recombinant 6 × His fusion proteins were expressed in E. coli BL21 (DE3) and purified to homogeneity using a Ni-NTA (Roche, USA) resin column. The oligonucleotide probes were amplified by the EMSA primers listed in Table S1 and labelled with biotin at the N′ end of the PCR product by the EMSA Probe Biotin Labelling Kit (Beyotime, China). The oligonucleotides containing only perfect elements were synthesized by Sangon Biotech (China) with 5′-biotin marked. EMSAs were performed as previously described (Li et al. 2018). Briefly, biotin-labelled probes were incubated with His-tagged OsHsfB4d protein at room temperature for 20 min, and the subsequent steps were performed using a Chemiluminescent EMSA Kit (Beyotime, China) according to the manufacturer’s instructions. Free and bound probes were separated via PAGE, transferred to nylon membranes and subjected to chemiluminescence examination.
ChIP-qPCR Assay
Leaves of 3-week-old pCXUN-Myc-OsHsfB4d seedlings were collected. Approximately 1 g leaves was harvested and cross-linked in 1% (v/v) formaldehyde at room temperature for 10 min, followed by neutralization with 0.125 M glycine. The chromatin-protein complex was isolated, resuspended in lysis buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% (w/v) SDS, 1% (v/v) Triton X-100, 1 mM PMSF, and 1% protease inhibitor mixture), and sheared by sonication to reduce the average DNA fragment size to 500 bp. Then, 50 μL of sheared chromatin was saved for use as an input control. ChIP was conducted using a ProFound™ c-Myc Tag IP/Co-IP kit according to the manufacturer’s instructions (Thermo Scientific, USA). The protein-DNA cross-links were reversed by incubating the immunoprecipitated complexes with 20 μL of 5 M NaCl at 65 °C overnight. The DNA was recovered by ethanol precipitation, washed with cold 75% ethanol and dried. The pellet was dissolved in water and analysed by qPCR. The enrichment of the target gene promoters is shown as the percentage of the input DNA. Actin was used as a nonspecific target gene. The primers used for qPCR are listed in Table S1.
Western-Blot Assay
Approximately 1 g of fresh rice leaves was ground in liquid nitrogen, resuspended in extraction buffer (50-mM Tris-HCl, pH 7.4, 150-mM NaCl, 5-mM EDTA, 1-mM PMSF, 1% NP-40 and 1× protease inhibitor cocktail) and then centrifuged at 12,000 g for 15 min at 4 °C. An aliquot of 100 μL of supernatant was boiled for 5 min with 1× SDS buffer. The protein samples were separated by SDS-PAGE using a 10% gel. Proteins were blotted onto a PVDF transfer membrane (Millipore, USA). OsHsfB4d protein was detected by a monoclonal anti-Myc antibody (Abcam, USA). The images were photographed using SuperSignal® West Dura Extended Duration Substrate (Thermo Scientific). The stained Rubisco protein shows the equal loading of protein samples.
Data Treatment
The accession number of the original transcription data of OsHsp18.0-CI is SRP079496 (Ju et al. 2017; Zuo et al. 2019). Quantitative data were analysed using Student’s t test (two-tailed t test with equal variances; Microsoft Excel) to evaluate the significance of the differences between wild-type plants and other plants. *Indicates significant (t test, P < 0.05) differences and **indicates P < 0.01.
Availability of Data and Materials
The data that support the findings of this study are available from the corresponding author on reasonable request.
Abbreviations
- BB:
-
Bacterial blight
- BLS:
-
Bacterial leaf streak
- DEG:
-
Differentially expressed genes
- dpi:
-
Days post inoculation
- DR:
-
Defense-related
- GFP:
-
Green fluorescent protein
- Hsfs:
-
Heat shock factors
- HSPs:
-
Heat shock proteins
- Luc:
-
Luciferase
- OE:
-
Overexpression
- PR:
-
Pathogenesis related
- QTLs:
-
Quantitative trait loci
- WT:
-
Wild-type
- Xoc:
-
Xanthomons oryzae pv. oryzicola
- Xoo:
-
Xanthomonas oryzae pv. oryzae
References
Baniwal SK, Bharti K, Chan KY, Fauth M, Ganguli A, Kotak S, Mishra SK, Nover L, Port M, Scharf KD, Tripp J, Weber C, Zielinski D, von Koskull-Doring P (2004) Heat stress response in plants, a complex game with chaperones and more than twenty heat stress transcription factors. J Biosci 29(4):471–487
Bechtold U, Albihlal WS, Lawson T, Fryer MJ, Sparrow PA, Richard F, Persad R, Bowden L, Hickman R, Martin C, Beynon JL, Buchanan-Wollaston V, Baker NR, Morison JIL, Schöffl F, Ott S, Mullineaux PM (2013) Arabidopsis HEAT SHOCK TRANSCRIPTION FACTORA1b overexpression enhances water productivity, resistance to drought, and infection. J Exp Bot 64(11):3467–3481
Bian XH, Li W, Niu CF, Wei W, Hu Y, Han JQ, Lu X, Tao JJ, Jin M, Qin H, Zhou B, Zhang WK, Ma B, Wang GD, Yu DY, Lai YC, Chen SY, Zhang JS (2020) A class B heat shock factor selected for during soybean domestication contributes to salt tolerance by promoting flavonoid biosynthesis. New Phytol 225(1):268–283
Chauhan H, Khurana N, Agarwal P, Khurana P (2011) Heat shock factors in rice (Oryza sativa L), genome-wide expression analysis during reproductive development and abiotic stress. Mol Gen Genomics 286(2):171
Chen S, Songkumarn P, Liu J, Wang G (2009) A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol 150(3):1111–1121
Evrard A, Kumar M, Lecourieux D, Lucks J, von Koskull-Döring P, Hirt H (2013) Regulation of the heat stress response in Arabidopsis by MPK6-targeted phosphorylation of the heat stress factor HsfA2. Peer J 1:e59
Feng C, Zhang X, Wu T, Yuan B, Ding X, Yao F, Chu Z (2016) The polygalacturonase-inhibiting protein 4 (OsPGIP4), a potential component of the qBlsr5a locus, confers resistance to bacterial leaf streak in rice. Planta 243(5):1297–1308
Fu J, Liu H, Li Y, Yu H, Li X, Xiao J, Wang S (2011) Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. Plant Physiol 155(1):589–602
Giesguth M, Sahm A, Simon S, Dietz KJ (2015) Redox-dependent translocation of the heat shock transcription factor AtHSFA8 from the cytosol to the nucleus in Arabidopsis thaliana. FEBS Lett 589(6):718–725
Guan JC, Jinn TL, Yeh CH, Feng SP, Chen YM, Lin CY (2004) Characterization of the genomic structures and selective expression profiles of nine class I small heat shock protein genes clustered on two chromosomes in rice (Oryza sativa L). Plant Mol Biol 56(5):795–809
Guan JC, Yeh CH, Lin YP, Ke YT, Chen MT, You JW, Liu YH, Lu CA, Wu SJ, Lin CY (2010) A 9 bp cis-element in the promoters of class I small heat shock protein genes on chromosome 3 in rice mediates L-azetidine-2-carboxylic acid and heat shock responses. J Exp Bot 61(15):4249–4261
Guo L, Guo C, Li M, Wang W, Luo C, Zhang Y, Chen L (2014) Suppression of expression of the putative receptor-like kinase gene NRRB enhances resistance to bacterial leaf streak in rice. Mol Boil Rep 41(4):2177–2187
Guo L, Li M, Wang W, Wang L, Hao G, Guo C, Chen L (2012) Over-expression in the nucleotide-binding site-leucine rich repeat gene DEPG1 increases susceptibility to bacterial leaf streak disease in transgenic rice plants. Mol Biol Rep 39(4):3491–3504
Hahn A, Bublak D, Schleiff E, Scharf KD (2011) Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 23(2):741–755
Hu Y, Han Y, Wei W, Li Y, Zhang K, Gao Y, Zhao FL, Feng J (2016) Identification, isolation, and expression analysis of heat shock transcription factors in the diploid woodland strawberry Fragaria vesca. Front Plant Sci 7:736
Hui S, Shi Y, Tian J, Wang L, Li Y, Wang S, Yuan M (2019) TALE-carrying bacterial pathogens trap host nuclear import receptors for facilitation of infection of rice. Mol Plant Pathol 20(4):519–532
Hutin M, Césari S, Chalvon V, Michel C, Tran TT, Boch J, Koebnik R, Szurek B, Kroj T (2016) Ectopic activation of the rice NLR heteropair RGA4/RGA5 confers resistance to bacterial blight and bacterial leaf streak diseases. Plant J 88(1):43–55
Ju Y, Tian H, Zhang R, Zuo L, Jin G, Xu Q, Ding X, Li X, Chu Z (2017) Overexpression of OsHSP180-CI enhances resistance to bacterial leaf streak in rice. Rice 10(1):12
Ke Y, Deng H, Wang S (2017) Advances in understanding broad-spectrum resistance to pathogens in rice. Plant J 90(4):738–748
Kotak S, Port M, Ganguli A, Bicker F, von Koskull-Döring P (2004) Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class a Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J 39(1):98–112
Kumar M, Busch W, Birke H, Kemmerling B, Nürnberger T, Schöffl F (2009) Heat shock factors HsfB1 and HsfB2b are involved in the regulation of PDF1.2 expression and pathogen resistance in Arabidopsis. Mol Plant 2(1):152–165
Lavania D, Dhingra A, Grover A (2018) Analysis of transactivation potential of rice (Oryza sativa L) heat shock factors. Planta 247(6):1267–1276
Li N, Chen J, Yang F, Wei S, Kong L, Ding X, Chu Z (2017) Identification of two novel Rhizoctonia solani-inducible cis-acting elements in the promoter of the maize gene, GRMZM2G315431. Sci Rep 7(1):42059
Li N, Kong L, Zhou W, Zhang X, Wei S, Ding X, Chu Z (2013) Overexpression of Os2H16 enhances resistance to phytopathogens and tolerance to drought stress in rice. Plant Cell Tissue Organ Cult 115(3):429–441
Li N, Wei S, Chen J, Yang F, Kong L, Chen C, Ding X, Chu Z (2018) OsASR2 regulates the expression of a defence-related gene, Os2H16, by targeting the GT-1 cis-element. Plant Biotechnol J 16(3):771–783
Lin YX, Jiang HY, Chu ZX, Tang XL, Zhu SW, Cheng BJ (2011) Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genomics 12(1):76
Liu HC, Charng YY (2013) Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol 163(1):276–290
Liu HC, Liao HT, Charng YY (2011) The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ 34(5):738–751
Ma H, Chen J, Zhang Z, Ma L, Yang Z, Zhan Q, Li X, Xiao J, Wang S (2017) MAPK kinase 102 promotes disease resistance and drought tolerance by activating different MAPKs in rice. Plant J 92(4):557–570
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu YG (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8(8):1274–1284
Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf KD (2002) In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev 16(12):1555–1567
Mittal D, Chakrabarti S, Sarkar A, Singh A, Grover A (2009) Heat shock factor gene family in rice, genomic organization and transcript expression profling in response to high temperature, low temperature and oxidative stresses. Plant Physiol Biochem 47(9):785–795
Mittal D, Enoki Y, Lavania D, Singh A, Sakurai H, Grover A (2011) Binding affinities and interactions among different heat shock element types and heat shock factors in rice (Oryza sativa L). FEBS J 278(17):3076–3085
Nelson R, Wiesner-Hanks T, Wisser R, Balint-Kurti P (2018) Navigating complexity to breed disease-resistant crops. Nat Rev Genet 19(1):21
Niño-Liu DO, Ronald PC, Bogdanove AJ (2006) Xanthomonas oryzae pathovars, model pathogens of a model crop. Mol Plant Pathol 7(5):303–324
Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD (2001) Arabidopsis and the heat stress transcription factor world, how many heat stress transcription factors do we need? Cell Stress Chaperon 6(3):177
Peng S, Zhu Z, Zhao K, Shi J, Yang Y, He M, Wang Y (2013) A Novel Heat Shock Transcription Factor, VpHsf1, from Chinese Wild Vitis pseudoreticulata is Involved in Biotic and Abiotic Stresses. Plant Mol Biol Rep 31(1):240–247.
Pick T, Jaskiewicz M, Peterhänsel C, Conrath U (2012) Heat shock factor HsfB1 primes gene transcription and systemic acquired resistance in Arabidopsis. Plant Physiol 159(1):52–55
Sakurai H, Enoki Y (2010) Novel aspects of heat shock factors, DNA recognition, chromatin modulation and gene expression. FEBS J 277(20):4140–4149
Scharf KD, Berberich T, Ebersberger I, Nover L (2012) The plant heat stress transcription factor (Hsf) family, structure, function and evolution. Biochim Biophys Acta 1819(2):104–119
Schramm F, Ganguli A, Kiehlmann E, Englich G, Walch D, von Koskull-Do Ring P (2006) The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol Biol 60(5):759–772
Sharma TR, Rai AK, Gupta SK, Vijayan J, Devanna BN, Ray S (2012) Rice blast management through host-plant resistance, retrospect and prospects. Agric Res 1(1):37–52
Shen X, Yuan B, Liu H, Li X, Xu C, Wang S (2010) Opposite functions of a rice mitogen-activated protein kinase during the process of resistance against Xanthomonas oryzae. Plant J 64(1):86–99
Tao Z, Liu H, Qiu D, Zhou Y, Li X, Xu C, Wang S (2009) A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol 151(2):936–948
Triplett LR, Cohen SP, Heffelfinger C, Schmidt CL, Huerta A, Tekete C, Verdier V, Bogdanove AJ, Leach JE (2016) A resistance locus in the American heirloom rice variety Carolina gold select is triggered by TAL effectors with diverse predicted targets and is effective against African strains of Xanthomonas oryzae pv oryzicola. Plant J 87(5):472–483
Waters ER (2012) The evolution, function, structure, and expression of the plant sHSPs. J Exp Bot 64(2):391–403
Wu T, Peng C, Li B, Wu W, Kong L, Li F, Chu Z, Liu F, Ding X (2019) OsPGIP1-mediated resistance to bacterial leaf streak in rice is beyond responsive to the polygalacturonase of Xanthomonas oryzae pv oryzicola. Rice 12(1):90
Xie X, Chen Z, Cao J, Guan H, Lin D, Li C, Lan T, Duan Y, Mao D, Wu W (2014) Toward the positional cloning of qBlsr5a, a QTL underlying resistance to bacterial leaf streak, using overlapping sub-CSSLs in rice. PLoS One 9(4):e95751
Xu M, Huang L, Zhang F, Zhu L, Zhou Y, Li Z (2013) Genome-wide phylogenetic analysis of stress-activated protein kinase genes in rice (OsSAPKs) and expression profiling in response to Xanthomonas oryzae pv Oryzicola infection. Plant Mol Biol Rep 31(4):877–885
Xu Y, Zhu XF, Zhou MG, Kuang J, Zhang Y, Shang Y, Wang JX (2010) Status of streptomycin resistance development in Xanthomonas oryzae pv oryzae and Xanthomonas oryzae pv oryzicola in China and their resistance characters. J Phytopathol 158(9):601–608
Yang F, Ding X, Chen J, Shen Y, Kong L, Li N, Chu Z (2017) Functional analysis of the GRMZM2G174449 promoter to identify Rhizoctonia solani-inducible cis-elements in maize. BMC Plant Biol 17(1):233
Yang W, Xu X, Li Y, Wang Y, Li M, Wang Y, Ding X, Chu Z (2016) Rutin-mediated priming of plant resistance to three bacterial pathogens initiating the early SA signal pathway. PLoS One 11(1):e0146910
Yang W, Zhang B, Qi G, Shang L, Liu H, Ding X, Chu Z (2019) Identification of the phytosulfokine receptor 1 (OsPSKR1) confers resistance to bacterial leaf streak in rice. Planta 250(5):1603–1612
Zhang R, Chen Z, Liu Y (2015) Advance in rice bacterial leaf streak researches. Agri Sci Tech 16(2):298–305
Zhang Y, Guo X, Cui Y, Guo C, Chen L (2017) Overexpression of the receptor-like cytoplasmic kinase gene XCRK enhances Xoc and oxidative stress tolerance in rice. J Plant Biol 60(5):523–532
Zhou J, Xu XC, Cao JJ, Yin LL, Xia XJ, Shi K, Zhou YH, Yu JQ (2018) Heat shock factor HsfA1a is essential for R gene-mediated nematode resistance and triggers H2O2 production. Plant Physiol 176(3):2456–2471
Zuo L, Zhang RH, Jin GX, Ju YH, Tian HJ, Ding XH, Chu ZH (2019) OsHsp180-CI regulates disease resistance to bacterial blight in rice. Acta Phytopathol Sin 49(1):90–100 (in Chinese with English abstract)
Acknowledgments
Not applicable.
Funding
This work was supported by the National Key Research and Development Program of China (2016YFD0100903), the National Natural Science Foundation of China (31771748, 31700219), and the Shandong Modern Agricultural Technology & Industry system (SDAIT-17-06).
Author information
Authors and Affiliations
Contributions
WY performed disease assessment, EMSA, ChIP-PCR, qRT-PCR, data analysis and drafted the manuscript; YJ and LZ performed the constructs, transgenic and disease assessment at T1 generations; XML helped in partial funds supporting and discussion, LS and XRL performed assistants in inoculation, vector construction, RNA extraction; SF helped in seed propagation; XD initiated and partially supervised the project; ZC supervised the project, designed the experiments, revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
The primers used in this study.
Additional file 2: Fig. S1.
Heatmaps showing the expression patterns of class A and B Hsfs in Hsp18.0-CI OE and WT plants post inoculation with Xoo or Xoc.
Additional file 3: Fig. S2.
Sequencing result for the fragment around the PAM in the wild type and the three OsHsfB4d-cas9 lines.
Additional file 4 Fig. S3
Nucleotide sequence of the promoter region of OsHsp18.0-CI. The translation start site of OsHsp18.0-CI is underlined with dots. The perfect HSE element is marked with light gray and the eight imperfect HSE are shown by dark gray. The probe 1 sequence is underlined in bold and the probe 2 sequence is indicated with wavy line, there is an overlap between the two sequences. Synthetic probe with only perfect HSE element is indicated by bold italic letters.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, W., Ju, Y., Zuo, L. et al. OsHsfB4d Binds the Promoter and Regulates the Expression of OsHsp18.0-CI to Resistant Against Xanthomonas Oryzae. Rice 13, 28 (2020). https://doi.org/10.1186/s12284-020-00388-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-020-00388-2