Abstract
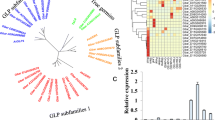
Fucosylation, one of the key posttranslational modifications, plays an important role in plants. It is involved in the development, signal transduction, reproduction, and disease resistance. α1,3-/4-Fucosyltransferase is responsible for transferring L-fucose from GDP-L-fucose to the N-glycan to exert fucosylational functions. However, the roles of the fucosyltransferase gene in cotton remain unknown. This study provided a comprehensive investigation of its possible functions. A genome-wide analysis identified four, four, eight, and eight FucT genes presented in the four sequenced cotton species, diploid Gossypium raimondii, G. arboreum, tetraploid G. hirsutum acc. TM-1, and G. barbadense cv. H7124, respectively. These FucTs were classified into two groups, with FucT4 homologs alone as a group. We isolated FucT4 in TM-1 and H7124, and named it GhFucT4 and GbFucT4, respectively. Quantitative RT-PCR and transcriptome data demonstrated that GhFucT4 had the highest expression levels in fibers among all GhFucT genes. Association studies and QTL co-localization supported the possible involvement of GhFucT4 in cotton fiber development. GhFucT4 and GbFucT4 shared high sequence identities, and FucT4 had higher expression in H7124 fiber tissues compared with TM-1. Furthermore, ectopic expression of FucT4 in transgenic Arabidopsis promoted root cell elongation, upregulated expression of genes related to cell wall loosening, and led to longer primary root. These results collectively indicate that FucT4 plays an important role in promoting cell elongation and modulating fiber development, which could be utilized to improve fiber quality traits in cotton breeding.





Similar content being viewed by others
Abbreviations
- DPA:
-
Days post-anthesis
- QTL:
-
Quantitative trait locus
- PAGE:
-
Polyacrylamide gel electrophoresis
- qRT-PCR:
-
Quantitative reverse transcription-polymerase chain reaction
- SNP:
-
Single nucleotide polymorphism
References
Bakker H, Schijlen E, de Vries T, Schiphorst WECM, Jordi W, Lommen A, Bosch D, van Die I (2001) Plant members of the α1→3/4-fucosyltransferase gene family encode an α1→4-fucosyltransferase, potentially involved in Lewisa biosynthesis, and two core α1→3-fucosyltransferases1. FEBS Lett 507:307–312
Bashline L, Lei L, Li S, Gu Y (2014) Cell wall, cytoskeleton, and cell expansion in higher plants. Mol Plant 7:586–600
Cai C, Ye W, Zhang T, Guo W (2014) Association analysis of fiber quality traits and exploration of elite alleles in Upland cotton cultivars/accessions (Gossypium hirsutum L.). J Integr Plant Biol 56:51–62
Campbell JA, Davies GJ, Bulone V, Henrissat B (1997) A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J 326(Pt 3):929–939
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cosgrove DJ (2000) Expansive growth of plant cell walls. Plant Physiol Biochem 38:109–124
Coutinho PM, Deleury E, Davies GJ, Henrissat B (2003) An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328:307–317
Du X, Huang G, He S, Yang Z, Sun G, Ma X, Li N, Zhang X, Sun J, Liu M, Jia Y, Pan Z, Gong W, Liu Z, Zhu H, Ma L, Liu F, Yang D, Wang F, Fan W, Gong Q, Peng Z, Wang L, Wang X, Xu S, Shang H, Lu C, Zheng H, Huang S, Lin T, Zhu Y, Li F (2018) Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat Genet 50:796–802
Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer ELL, Bateman A (2006) Pfam: clans, web tools and services. Nucleic Acids Res 34:D247–D251
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37
Fry SC, Aldington S, Hetherington PR, Aitken J (1993) Oligosaccharides as signals and substrates in the plant cell wall. Plant Physiol 103:1–5
Fu Y, Yuan D, Hu W, Cai C, Guo W (2013) Development of Gossypium barbadense chromosome 18 segment substitution lines in the genetic standard line TM-1 of Gossypium hirsutum and mapping of QTLs related to agronomic traits. Acta Agronomica Sinica 39:21
Graves DA, Stewart JM (1988) Chronology of the differentiation of cotton (Gossypium hirsutum L.) fiber cells. Planta 175:254–258
Guo X, Runavot JL, Bourot S, Meulewaeter F, Hernandez-Gomez M, Holland C, Harholt J, Willats WGT, Mravec J, Knox P, Ulvskov P (2019) Metabolism of polysaccharides in dynamic middle lamellae during cotton fibre development. Planta 249:1565–1581
Haigler CH, Betancur L, Stiff MR, Tuttle JR (2012) Cotton fiber: a powerful single-cell model for cell wall and cellulose research. Front Plant Sci 3:104
Harmoko R, Yoo JY, Ko KS, Ramasamy NK, Hwang BY, Lee EJ, Kim Ho S, Lee KJ, Oh D-B, Kim D-Y, Lee S, Li Y, Lee SY, Lee KO (2016) N-glycan containing a core α1,3-fucose residue is required for basipetal auxin transport and gravitropic response in rice (Oryza sativa). New Phytol 212:108–122
Howe EA, Sinha R, Schlauch D, Quackenbush J (2011) RNA-Seq analysis in MeV. Bioinformatics 27:3209–3210
Hu Y, Chen J, Fang L, Zhang Z, Ma W, Niu Y, Ju L, Deng J, Zhao T, Lian J, Baruch K, Fang D, Liu X, Ruan YL, Rahman MU, Han J, Wang K, Wang Q, Wu H, Mei G, Zang Y, Han Z, Xu C, Shen W, Yang D, Si Z, Dai F, Zou L, Huang F, Bai Y, Zhang Y, Brodt A, Ben-Hamo H, Zhu X, Zhou B, Guan X, Zhu S, Chen X, Zhang T (2019) Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet 51:739–748
Jansing J, Sack M, Augustine SM, Fischer R, Bortesi L (2019) CRISPR/Cas9-mediated knockout of six glycosyltransferase genes in Nicotiana benthamiana for the production of recombinant proteins lacking beta-1,2-xylose and core alpha-1,3-fucose. Plant Biotechnol J 17:350–361
Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, Soltis DE, Clifton SW, Schlarbaum SE, Schuster SC, Ma H, Leebens-Mack J, dePamphilis CW (2011) Ancestral polyploidy in seed plants and angiosperms. Nature 473:97–100
Kohel RJ, Richmond TR, Lewis CF (1970) Texas Marker-1. Description of a genetic standard for Gossypium hirsutum L.1. Crop Sci 10:670–671
Lee J, Burns TH, Light G, Sun Y, Fokar M, Kasukabe Y, Fujisawa K, Maekawa Y, Allen RD (2010) Xyloglucan endotransglycosylase/hydrolase genes in cotton and their role in fiber elongation. Planta 232:1191–1205
Levy S, York WS, Stuikeprill R, Meyer B, Staehelin LA (1991) Simulations of the static and dynamic molecular-conformations of xyloglucan—the role of the fucosylated side-Chain in surface-specific side-chain folding. Plant J 1:195–215
Li Y, Tu L, Pettolino FA, Ji S, Hao J, Yuan D, Deng F, Tan J, Hu H, Wang Q, Llewellyn DJ, Zhang X (2016) GbEXPATR, a species-specific expansin, enhances cotton fibre elongation through cell wall restructuring. Plant Biotechnol J 14:951–963
Liang W, Fang L, Xiang D, Hu Y, Feng H, Chang L, Zhang T (2015) Transcriptome analysis of short fiber mutant ligon lintless-1 (Li1) reveals critical genes and key pathways in cotton fiber elongation and leaf development. PLoS ONE 10:e0143503
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Lv F, Li P, Zhang R, Li N, Guo W (2016) Functional divergence of GhCFE5 homoeologs revealed in cotton fiber and Arabidopsis root cell development. Plant Cell Rep 35:867–881
Ma B, Simala-Grant JL, Taylor DE (2006) Fucosylation in prokaryotes and eukaryotes. Glycobiology 16:158R–184R
McDougall GJ, Fry SC (1989) Structure-activity relationships for xyloglucan oligosaccharides with antiauxin activity. Plant Physiol 89:883–887
Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, Llewellyn D, Showmaker KC, Shu S, Udall J, Yoo MJ, Byers R, Chen W, Doron-Faigenboim A, Duke MV, Gong L, Grimwood J, Grover C, Grupp K, Hu G, Lee TH, Li J, Lin L, Liu T, Marler BS, Page JT, Roberts AW, Romanel E, Sanders WS, Szadkowski E, Tan X, Tang H, Xu C, Wang J, Wang Z, Zhang D, Zhang L, Ashrafi H, Bedon F, Bowers JE, Brubaker CL, Chee PW, Das S, Gingle AR, Haigler CH, Harker D, Hoffmann LV, Hovav R, Jones DC, Lemke C, Mansoor S, ur Rahman M, Rainville LN, Rambani A, Reddy UK, Rong JK, Saranga Y, Scheffler BE, Scheffler JA, Stelly DM, Triplett BA, Van Deynze A, Vaslin MF, Waghmare VN, Walford SA, Wright RJ, Zaki EA, Zhang T, Dennis ES, Mayer KF, Peterson DG, Rokhsar DS, Wang X, Schmutz J (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492:423–427
Qin YM, Zhu YX (2011) How cotton fibers elongate: a tale of linear cell-growth mode. Curr Opin Plant Biol 14:106–111
Rautengarten C, Ebert B, Liu L, Stonebloom S, Smith-Moritz AM, Pauly M, Orellana A, Scheller HV, Heazlewood JL (2016) The Arabidopsis Golgi-localized GDP-L-fucose transporter is required for plant development. Nat Commun 7:12119
Shang X, Chai Q, Zhang Q, Jiang J, Zhang T, Guo W, Ruan Y-L (2015) Down-regulation of the cotton endo-1,4-beta-glucanase gene KOR1 disrupts endosperm cellularization, delays embryo development, and reduces early seedling vigour. J Exp Bot 66:3071–3083
Shen X, Guo W, Zhu X, Yuan Y, Yu JZ, Kohel RJ, Zhang T (2005) Molecular mapping of QTLs for fiber qualities in three diverse lines in Upland cotton using SSR markers. Mol Breeding 15:169–181
Sim JS, Kesawat MS, Kumar M, Kim SY, Mani V, Subramanian P, Park S, Lee CM, Kim SR, Hahn BS (2018) Lack of the alpha1,3-Fucosyltransferase gene (Osfuct) affects anther development and pollen viability in rice. Int J Mol Sci 19(4):1225
Singh B, Avci U, Inwood SEE, Grimson MJ, Landgraf J, Mohnen D, Sorensen I, Wilkerson CG, Willats WGT, Haigler CH (2009) A specialized outer layer of the primary cell wall joins elongating cotton fibers into tissue-like bundles. Plant Physiol 150:684–699
Strasser R, Altmann F, Mach L, Glossl J, Steinkellner H (2004) Generation of Arabidopsis thaliana plants with complex N-glycans lacking beta 1,2-linked xylose and core alpha 1,3-linked fucose. FEBS Lett 561:132–136
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Wang H, Guo Y, Lv F, Zhu H, Wu S, Jiang Y, Li F, Zhou B, Guo W, Zhang T (2010) The essential role of GhPEL gene, encoding a pectate lyase, in cell wall loosening by depolymerization of the de-esterified pectin during fiber elongation in cotton. Plant Mol Biol 72:397–406
Wang W, Zhang X, Niittylä T (2019a) OPENER is a nuclear envelope and mitochondria localized protein required for cell cycle progression in Arabidopsis. Plant Cell 31:1446–1465
Wang Z, Yang Z, Li F (2019b) Updates on molecular mechanisms in the development of branched trichome in Arabidopsis and nonbranched in cotton. Plant Biotechnol J 17:1706–1722
Wilson IB, Rendic D, Freilinger A, Dumic J, Altmann F, Mucha J, Muller S, Hauser MT (2001) Cloning and expression of cDNAs encoding alpha1,3-fucosyltransferase homologues from Arabidopsis thaliana. Biochim Biophys Acta 1527:88–96
Wilson MH, Holman TJ, Sorensen I, Cancho-Sanchez E, Wells DM, Swarup R, Knox JP, Willats WG, Ubeda-Tomas S, Holdsworth M, Bennett MJ, Vissenberg K, Hodgman TC (2015) Multi-omics analysis identifies genes mediating the extension of cell walls in the Arabidopsis thaliana root elongation zone. Front Cell Dev Biol 3:10
Xu W, Zhang D, Wu Y, Qin L, Huang G, Li J, Li L, Li X (2013) Cotton PRP5 gene encoding a proline-rich protein is involved in fiber development. Plant Mol Biol 82:353–365
Xu P, Cao Z, Zhang X, Gao J, Zhang X, Shen X (2014) Identification of quantitative trait loci for fiber quality properties on homoeologous chromosomes 13 and 18 of Gossypium klotzschianum. Crop Sci 54:484–491
Zablackis E, York WS, Pauly M, Hantus S, Reiter WD, Chapple CCS, Albersheim P, Darvill A (1996) Substitution of L-fucose by L-galactose in cell walls of Arabidopsis mur1. Science 272:1808–1810
Acknowledgements
We are grateful to Dr. Xiongming Du, Institute of Cotton Research of Chinese Academy of Agricultural Sciences, for providing the phenotypic data of the natural population consisting of 278 G. hirsutum cultivars. Many thanks to Miss Yue Feng in Nanjing Agricultural University for the preparation of preliminary plant materials.
Funding
This work was supported by the National Natural Science Foundation of China (31701472), the Fundamental Research Funds for the Central Universities (KJQN201801, KJQN201806), and Jiangsu Collaborative Innovation Center for Modern Crop Production project (No. 10). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
GWZ conceived the study, GWZ and SXG designed the experiments, SXG, ZLJ, and DYJ performed the experiments, SXG analyzed the data and drafted the manuscript, and GWZ revised the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.

438_2020_1687_MOESM1_ESM.jpg
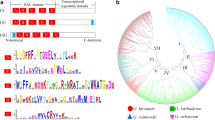
Supplementary file1 Fig. S1 Sequence analysis of GhFucT gene. a Phylogenetic analysis of isolated GhFucT genes and Arabidopsis thaliana FucT family genes. b Sequence alignment of GhFucT and AtFucT_C amino acids. c Analysis of deduced GhFucT protein. The signal peptide in red, Glyco_transf_10 domain in blue, glycosylation amino acid sites in green and phosphorylation amino acid sites in blue were indicated and drawn with IBS software (JPG 2693 kb)


438_2020_1687_MOESM3_ESM.jpg
Supplementary file3 Fig. S3 Hypocotyl length between wild type and transgenic Arabidopsis etiolated seedlings. Average hypocotyl lengths of WT and transgenic etiolated seedlings after 4 days growth under darkness conditions. Significant differences between wild type and transgenic lines were calculated by the one-way analysis of variance (ANOVA) test (p < 0.05). Vertical bars represented standard error (SE) (JPG 252 kb)
Rights and permissions
About this article
Cite this article
Shang, X., Zhu, L., Duan, Y. et al. A cotton α1,3-/4-fucosyltransferase-encoding gene, FucT4, plays an important role in cell elongation and is significantly associated with fiber quality. Mol Genet Genomics 295, 1141–1153 (2020). https://doi.org/10.1007/s00438-020-01687-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-020-01687-5