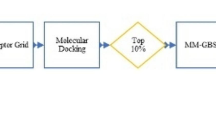
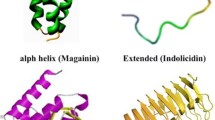
Abstract
Multi-drug resistance is a major issue faced by the global pharmaceutical industry. Short antimicrobial peptides such as anoplins can be used to replace antibiotics, thus mitigating this issue. Antimicrobial activity, non-toxicity, and structural stability are essential features of a therapeutic drug. Antimicrobial activity and toxicity to human erythrocytes have been previously reported for anoplin and anoplin R5K T8W. This study attempts to identify a therapeutic peptide drug scaffold between these peptides by examining their structural stability, mainly based on the hydrogen bonds (H-bond) found in their structures. The static structure of anoplin R5K T8W displayed lower H-bond distances than anoplin, thereby exhibiting enhanced structural stability. Dynamic stability studies revealed that conformers of anoplin R5K T8W exhibited lower hydrogen bond distances (HBDs), higher H-bond occupancies, and higher radial distribution function (RDF) of H-bonds in comparison with conformers of anoplin. Furthermore, conformers of anoplin R5K T8W generated using 50-ns molecular dynamics simulation displayed lower conformational free energy than anoplin, thus establishing its higher structural stability. Overall, anoplin R5K T8W can be claimed as a promising scaffold that may be used for therapeutic purposes. In conclusion, H-bonds play a major role in structural stability and may aid in identification of a therapeutic peptide scaffold.

Graphical abstract






Similar content being viewed by others
References
Jenssen H, Hamill P, Hancock RE (2006) Peptide antimicrobial agents. Clin Microbiol Rev 9:491–511. https://doi.org/10.1128/CMR.00056-05
Phambu N, Almarwani B, Garcia AM, Hamza NS, Muhsen A, Baidoo JE, Sunda-Meya A (2017) Chain length effect on the structure and stability of antimicrobial peptides of the (RW) n series. Biophys Chem 227:8–13. https://doi.org/10.1016/j.bpc.2017.05.009
Seo MD, Won HS, Kim JH, Mishig-Ochir T, Lee BJ (2012) Antimicrobial peptides for therapeutic applications: a review. Molecules 17:12276–12286. https://doi.org/10.3390/molecules171012276
Wang S, Zeng X, Yang Q, Qiao S (2016) Antimicrobial peptides as potential alternatives to antibiotics in food animal industry. Int J Mol Sci 17:603. https://doi.org/10.3390/ijms17050603
Rausch JM, Marks JR, Rathinakumar R, Wimley WC (2007) β-Sheet pore-forming peptides selected from a rational combinatorial library: mechanism of pore formation in lipid vesicles and activity in biological membranes. Biochemistry 46:12124–12139. https://doi.org/10.1021/bi700978h
Giuliani A, Pirri G, Nicoletto S (2007) Antimicrobial peptides: an overview of a promising class of therapeutics. Open Life Sci 2:1–33. https://doi.org/10.2478/s11535-007-0010-5
Herbel V, Wink M (2016) Mode of action and membrane specificity of the antimicrobial peptide snakin-2. PeerJ 4:e1987. https://doi.org/10.7717/peerj.1987
Dos Santos Cabrera MP, Arcisio-Miranda M, Broggio Costa ST, Konno K, Ruggiero JR, Procopio J, Ruggiero Neto J (2008) Study of the mechanism of action of anoplin, a helical antimicrobial decapeptide with ion channel-like activity, and the role of the amidated C-terminus. J Pept Sci 14:661–669. https://doi.org/10.1002/psc.960
Fair RJ, Tor Y (2014) Antibiotics and bacterial resistance in the 21st century. Perspect Med Chem 6:PMC-S14459. https://doi.org/10.4137/PMC.S14459
Sengupta S, Chattopadhyay MK, Grossart HP (2013) The multifaceted roles of antibiotics and antibiotic resistance in nature. Front Microbiol 4:47. https://doi.org/10.3389/fmicb.2013.00047
Kang SJ, Park SJ, Mishig-Ochir T, Lee BJ (2014) Antimicrobial peptides: therapeutic potentials. Expert Rev Anti-Infect Ther 12:1477–1486. https://doi.org/10.1586/14787210.2014.976613
Mahlapuu M, Håkansson J, Ringstad L, Björn C (2016) Antimicrobial peptides: an emerging category of therapeutic agents. Front Cell Infect Microbiol 6:194. https://doi.org/10.3389/fcimb.2016.00194
Da Costa JP, Cova M, Ferreira R, Vitorino R (2015) Antimicrobial peptides: an alternative for innovative medicines? Appl Microbiol Biotechnol 99:2023–2040. https://doi.org/10.1007/s00253-015-6375-x
Lau L, Dunn MK (2018) Therapeutic peptides: historical perspectives, current development trends, and future directions. Bioorg Med Chem 26:2700–2707. https://doi.org/10.1016/j.bmc.2017.06.052
Pennington MW, Czerwinski A, Norton RS (2018) Peptide therapeutics from venom: current status and potential. Bioorg Med Chem 26:2738–2758. https://doi.org/10.1016/j.bmc.2017.09.029
Munk JK, Ritz C, Fliedner FP, Frimodt-Møller N, Hansen PR (2014) Novel method to identify the optimal antimicrobial peptide in a combination matrix, using anoplin as an example. Antimicrob Agents Chemother 58:1063–1070. https://doi.org/10.1128/AAC.02369-13
Konno K, Hisada M, Fontana R, Lorenzi CC, Naoki H, Itagaki Y, Miwa A, Kawai N, Nakata Y, Yasuhara T, Neto JR (2001) Anoplin, a novel antimicrobial peptide from the venom of the solitary wasp Anoplius samariensis. Biochim Biophys Acta Protein Struct Mol Enzymol 1550:70–80. https://doi.org/10.1016/S0167-4838(01)00271-0
Munk JK, Uggerhøj LE, Poulsen TJ, Frimodt-Møller N, Wimmer R, Nyberg NT, Hansen PR (2013) Synthetic analogs of anoplin show improved antimicrobial activities. J Pept Sci 19:669–675. https://doi.org/10.1002/psc.2548
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242. https://doi.org/10.1093/nar/28.1.235
Uggerhøj LE, Poulsen TJ, Munk JK, Fredborg M, Sondergaard TE, Frimodt-Moller N, Hansen PR, Wimmer R (2015) Rational design of alpha-helical antimicrobial peptides: do’s and don’ts. ChemBioChem 16:242–253. https://doi.org/10.1002/cbic.201402581
Wang Y, Chen J, Zheng X, Yang X, Ma P, Cai Y, Zhang B, Chen Y (2014) Design of novel analogs of short antimicrobial peptide anoplin with improved antimicrobial activity. J Pept Sci 20:945–951. https://doi.org/10.1002/psc.2705
Krieger E YASARA Version 16.11.20. Bioinformatics 30 (© 1993–2016):2891–2892
Krieger E, Darden T, Nabuurs SB, Finkelstein A, Vriend G (2004) Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins Struct Funct Bioinf 57:678–683. https://doi.org/10.1002/prot.20251
Baker EN, Hubbard RE (1984) Hydrogen bonding in globular proteins. Prog Biophys Mol Biol 44:97–179. https://doi.org/10.1016/0079-6107(84)90007-5
Mills JEJ, Dean PM (1996) Three-dimensional hydrogen-bond geometry and probability information from a crystal survey. J Comput Aided Mol Des 10:607–622. https://doi.org/10.1007/BF00134183
Remer LC, Jensen JH (2000) Toward a general theory of hydrogen bonding: the short, strong hydrogen bond [HOH… OH]. J Phys Chem A 104:9266–9275. https://doi.org/10.1021/jp002726n
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF chimera--a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Saravanan KM, Selvaraj S (2017) Dihedral angle preferences of amino acid residues forming various non-local interactions in proteins. J Biol Phys 43:265–278. https://doi.org/10.1007/s10867-017-9451-x
Heinig M, Frishman D (2004) STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res 32:W500–W502. https://doi.org/10.1093/nar/gkh429
Sforça ML, Oyama S, Canduri F, Lorenzi CC, Pertinhez TA, Konno K, Souza BM, Palma MS, Ruggiero Neto J, Azevedo WF, Spisni A (2004) How C-terminal carboxyamidation alters the biological activity of peptides from the venom of the eumenine solitary wasp. Biochemistry 43:5608–5617. https://doi.org/10.1021/bi0360915
Mura M, Wang J, Zhou Y, Pinna M, Zvelindovsky AV, Dennison SR, Phoenix DA (2016) The effect of amidation on the behaviour of antimicrobial peptides. Eur Biophys J 45:195–207. https://doi.org/10.1007/s00249-015-1094-x
Microsoft ® Word ® for Office 365 MSO (16.0.11425.20200), Version 1903, 32-bit, OFFICE 2016 DESKTOP
Paint, Microsoft Windows, Version 1809, (OS Build 17763.404), © 2018 Microsoft Corporation
Seeliger D, Haas J, De Groot BL (2007) Geometry-based sampling of conformational transitions in proteins. Structure 15:1482–1492. https://doi.org/10.1016/j.str.2007.09.017
Bekker H, Berendsen HJC, Dijkstra EJ, Achterop S, van Drunen R, van der Spoel D, Sijbers A, Keegstra H, Reitsma B, Renardus MKR (1993) Gromacs: a parallel computer for molecular dynamics simulations. In: de Groot RA, Nadrchal J (eds.) Physics Computing 92. World Scientific 252-256
Berendsen HJC, van der Spoel D, van Drunen R (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun 91:43–56. https://doi.org/10.1016/0010-4655(95)00042-E
Lindahl E, Hess B, van der Spoel D (2001) Gromacs 3.0: a package for molecular simulation and trajectory analysis. J Mol Model 7:306–317. https://doi.org/10.1007/s008940100045
van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC (2005) GROMACS: fast, flexible and free. J Comput Chem 26:1701–1718. https://doi.org/10.1002/jcc.20291
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38. https://doi.org/10.1016/0263-7855(96)00018-5
Pedretti A, Villa L, Vistoli G (2004) VEGA–an open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming. J Comput Aided Mol Des 18:167–173. https://doi.org/10.1023/B:JCAM.0000035186.90683.f2
Microsoft ® Excel ® for Office 365 MSO (16.0.11425.20200), Version 1903, 32-bit, OFFICE 2016 DESKTOP
Chen J, Wang J, Zhang Q, Chen K, Zhu W (2015) A comparative study of trypsin specificity based on QM/MM molecular dynamics simulation and QM/MM GBSA calculation. J Biomol Struct Dyn 33:2606–2618. https://doi.org/10.1080/07391102.2014.1003146
Azam SS, Abro A, Raza S (2015) Binding pattern analysis and structural insight into the inhibition mechanism of sterol 24-C methyltransferase by docking and molecular dynamics approach. J Biomol Struct Dyn 33:2563–2577. https://doi.org/10.1080/07391102.2014.1002423
StatPlus, AnalystSoft Inc.-statistical analysis program. Version v6. See http://www.analystsoft.com/en/
Vadivel K, Namasivayam G (2009) An estimate of the numbers and density of low-energy structures (or decoys) in the conformational landscape of proteins. PLoS One 4:e5418. https://doi.org/10.1371/journal.pone.0005148
Liu S, Zhang C, Zhou H, Zhou Y (2004) A physical reference state unifies the structure-derived potential of mean force for protein folding and binding. Proteins 56:93–101. https://doi.org/10.1002/prot.20019
Zhou H, Zhou Y (2002) Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci 11:2714–2726. https://doi.org/10.1110/ps.0217002
Kusunoki H, MacDonald RI, Mondragón A (2004) Structural insights into the stability and flexibility of unusual erythroid spectrin repeats. Structure. 12:645–656. https://doi.org/10.1016/j.str.2004.02.022
Acknowledgments
The authors thank Vellore Institute of Technology (Deemed to be University) for providing “VIT SEED GRANT” for carrying out this research work.
Data availability statement
Data other than supplementary data will be made available on request.
Author information
Authors and Affiliations
Contributions
Conceptualization: Shruti Sunil Ranade; methodology: Shruti Sunil Ranade; formal analysis and investigation: Shruti Sunil Ranade; writing—original draft preparation: Shruti Sunil Ranade; writing—review and editing: Rajasekaran Ramalingam; funding acquisition: Rajasekaran Ramalingam; resources: Rajasekaran Ramalingam; supervision: Rajasekaran Ramalingam.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ranade, S.S., Ramalingam, R. Hydrogen bonds in anoplin peptides aid in identification of a structurally stable therapeutic drug scaffold. J Mol Model 26, 155 (2020). https://doi.org/10.1007/s00894-020-04380-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-020-04380-x