Abstract
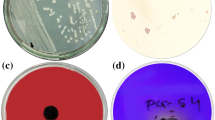
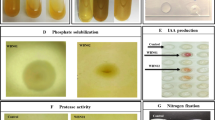
In agriculture, instead of synthetic fertilizers, natural bio-inoculants can be used to increase growth and yield of crops. For this purpose, we report a thermophilic bacteria Klebsiella sp. strain PMnew, isolated from Paniphala hot spring. The strain was characterized and assessed for plant growth-promoting traits. Oryza sativa L. var Swarna (rice) seeds were inoculated with the strain to study the bacterization effect on vegetative and reproductive growth of rice plants. The results indicate that PMnew produces organic acids to solubilize phosphate (550.16 ± 0.04 µg/ml), fixes nitrogen, produces indole compounds, siderophore, and ACC deaminase, and shows heavy metal resistance to chromium, cobalt, arsenic, cadmium, and mercury. It also possesses the ability to utilize several monomeric and polymeric sugars as sole carbon source including starch, agar, xylan, gelatin, and pectin, and can grow under both nutrient-rich and deficient conditions. Inoculated rice plants grew twice the length of control plants and surpassed the total grain mass yield of control plants by almost 18 times. Thus, this study brings forth a broad spectrum and easy to cultivate bio-inoculant, which can be used to increase rice production.




Similar content being viewed by others
References
Savci S (2012) Investigation of effect of chemical fertilizers on environment. Apcbee Procedia 1:287–292
Singh S, Singh BK, Yadav SM, Gupta AK (2014) Potential of biofertilizers in crop production in Indian agriculture. Am J Plant Nutr Fertil Technol 4:33–40
Ahemad M, Kibret M (2014) Mechanisms and applications of plant growth promoting rhizobacteria: current perspective. J King Saud Univ Sci 26:1–20
Numan M, Bashir S, Khan Y et al (2018) Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: a review. Microbiol Res 209:21–32
Burd GI, Dixon DG, Glick BR (2000) Plant growth-promoting bacteria that decrease heavy metal toxicity in plants. Can J Microbiol 46:237–245
Singh RP, Jha P, Jha PN (2015) The plant-growth-promoting bacterium Klebsiella sp. SBP-8 confers induced systemic tolerance in wheat (Triticum aestivum) under salt stress. J Plant Physiol 184:57–67
Penrose DM, Glick BR (2003) Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plant 118:10–15
Banik A, Mukhopadhaya SK, Dangar TK (2016) Characterization of N2-fixing plant growth promoting endophytic and epiphytic bacterial community of Indian cultivated and wild rice (Oryza spp.) genotypes. Planta 243:799–812
Stewart WDP (1970) Nitrogen fixation by blue-green algae in Yellowstone thermal areas. Phycologia 9:261–268
Verma JP, Jaiswal DK, Krishna R et al (2018) Characterization and screening of thermophilic bacillus strains for developing plant growth promoting consortium from hot spring of Leh and Ladakh Region of India. Front Microbiol 9:1293
Absher M (1973) Hemocytometer counting. Tissue Cult 1:395–397
Strober W (2001) Trypan blue exclusion test of cell viability. In: Current protocols in immunology. Wiley, Hoboken, pp A.3B.1–A.3B.2
Stolp H, Gadkari D (1981) Non-pathogenic members of the genus Pseudomonas. In: Mortimer PS, Heinz S, Trüper HG, et al. (eds) The prokaryotes: a handbook on habitats, isolation, and identification of bacteria. Springer, New York, pp 1302–1330
Smibert R, Krieg N (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg N (eds) Methods for general and molecular bacteriology. Washington DC, pp 607–654
Bauer AW, Kirby WM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45:493–496
Innis MA, Gelfand D, Sninsky JJ, White TJ (1990) PCR protocols: a guide to methods and applications. Academic Press, Cambridge
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Rice P, Longden I, Bleasby A (2000) EMBOSS: the European molecular biology open software suite. Trends Genet 16:276–277
Rahman F, Meryandini A, Junior M, Rusmana I (2011) Isolation and identification of an agar-liquefying marine bacterium and some properties of its extracellular agarases. Biodiversitas 10:192–197
Balan SS, Nethaji R, Sankar S, Jayalakshmi S (2012) Production of gelatinase enzyme from Bacillus spp. isolated from the sediment sample of Porto Novo Coastal sites. Asian Pac J Trop Biomed 2:S1811–S1816
Tran LH, Nagano H (2006) Isolation and characteristics of Bacillus subtilis CN2 and its collagenase production. J Food Sci 67:1184–1187
Bailey MJ, Biely P, Poutanen K (1992) Interlaboratory testing of methods for assay of xylanase activity. J Biotechnol 23:257–270
Anisa SK, Ashwini S, Girish K (2013) Isolation and screening of Aspergillus spp. for pectinolytic activity. Electron J Biol 9:37–41
Tao Y, Jin H, Long Z et al (2005) Purification and characterization of an extracellular chitinase produced by bacterium C4. Ann Microbiol 55:213–218
Kumar M, Rana S, Beniwal V, Salar RK (2015) Optimization of tannase production by a novel Klebsiella pneumoniae KP715242 using central composite design. Biotechnol Rep 7:128–134
von Tigerstrom RG, Stelmaschuk S (1989) The use of Tween 20 in a sensitive turbidimetric assay of lipolytic enzymes. Can J Microbiol 35:511–514
Hardy RWF, Holsten RD, Jackson EK, Burns RC (1968) The acetylene-ethylene assay for N2 fixation: laboratory and field evaluation. Plant Physiol 43:1185–1207
Cappuccino JG, Sherman N (2014) Biochemical activities of microorganisms. In: Microbiology : a laboratory manual. Pearson, New York, p 544
Murphy J, Riley JP (1962) A modified single solution method for the determination of phosphate in natural waters. Anal Chim Acta 27:31–36
Wang Z, Xu G, Ma P et al (2017) Isolation and characterization of a phosphorus-solubilizing bacterium from rhizosphere soils and its colonization of chinese cabbage (Brassica campestris ssp. chinensis). Front Microbiol 8:1270
Mitchell JW, Brunstetter BC (1939) Colorimetric methods for the quantitative estimation of indole(3) acetic acid. Bot Gaz 100:802–816
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Daware V, Patil RS, Gade WN (2015) Isolation, identification and characterization of heavy metal tolerant bacteria from Mula river, Pune, Maharashtra, India. Trends Biotechnol Res 4:1–10
Lee C-H, Liu J-W, Su L-H et al (2010) Hypermucoviscosity associated with Klebsiella pneumoniae-mediated invasive syndrome: a prospective cross-sectional study in Taiwan. Int J Infect Dis 14:e688–e692
Aher T, Roy A, Kumar P (2012) Molecular detection of virulence genes associated with pathogenicity of Klebsiella spp. isolated from the respiratory tract of apparently healthy as well as sick goats. Isr J Vet Med 67:249–252
Kahn D, Hawkins M, Eady R (1982) Nitrogen fixation in Klebsiella pneumoniae: nitrogenase levels and the effect of added molybdate on nitrogenase derepressed under molybdenum deprivation ProDom View project. J Gen Microbiol 128:779–787
Sachdev DP, Chaudhari HG, Kasture VM et al (2009) Isolation and characterization of indole acetic acid (IAA) producing Klebsiella pneumoniae strains from rhizosphere of wheat (Triticum aestivum) and their effect on plant growth. Indian J Exp Biol 47:993–1000
Rönkko R, Laakso T, Williams P, Korhonen T (1987) Root-associated Enterobacter and Klebsiella in Poa pratensis: characterization of an iron-scavenging system and a substance stimulating root hair production. Mol Plant-Microbe Interact 3:358–365
Brisse S, Grimont F, Grimont PAD (2006) The genus Klebsiella. The prokaryotes. Springer, New York, pp 159–196
Liu W, Wang Q, Hou J et al (2016) Whole genome analysis of halotolerant and alkalotolerant plant growth-promoting rhizobacterium Klebsiella sp. D5A. Sci Rep 6:26710
El-Khawas H, Adachi K (1999) Identification and quantification of auxins in culture media of Azospirillum and Klebsiella and their effect on rice roots. Biol Fertil Soils 28:377–381
Kumar A, Chakraborti S, Joshi P et al (2011) A multiple antibiotic and serum resistant oligotrophic strain, Klebsiella pneumoniae MB45 having novel dfrA30, is sensitive to ZnO QDs. Ann Clin Microbiol Antimicrob 10:19
Deschamps AM, Comtat J, Nouvion N, Lebeault JM (1982) Degradation of purified birch-wood xylan and production of xylanase by wood-decaying bacteria. J Gen Appl Microbiol 28:275–280
Yasin AR, Salman SA, Al-Mayaly Ikamila I (2014) Bioremediation of polluted water with crude oil in South Baghdad power plant. Iraqi J Sci 55:113–122
Gawande BN, Patkar AY (2001) Purification and properties of a novel raw starch degrading-cyclodextrin glycosyltransferase from Klebsiella pneumoniae AS-22. Enzyme Microb Technol 28:735–743
Anand AAP, Vennison SJ, Sankar SG et al (2010) Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J Insect Sci 10:107
Song T, Zhang W, Wei C et al (2015) Isolation and characterization of agar-degrading endophytic bacteria from plants. Curr Microbiol 70:275–281
Baron AJ, Wong TY, Hicks SJ et al (1994) Alginate lyase from Klebsiella pneumoniae, subsp. aerogenes: gene cloning, sequence analysis and high-level production in Escherichia coli. Gene 143:61–66
Acknowledgement
This study was funded by DST-INSPIRE (Innovation in Scientific Pursuit for Inspired Research, Department of Science and Technology) (IF 140017), Ministry of Science and Technology, Govt. of India, and UGC-sponsored Major Research Project Grant (MRP-MAJOR-MICR-2013-7783).
Author information
Authors and Affiliations
Contributions
TM, AB, and SKM conceived and designed the research. TM, AB, and SKM conducted the experiments and analyzed the data. TM, AB, and SKM wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mukherjee, T., Banik, A. & Mukhopadhyay, S.K. Plant Growth-Promoting Traits of a Thermophilic Strain of the Klebsiella Group with its Effect on Rice Plant Growth. Curr Microbiol 77, 2613–2622 (2020). https://doi.org/10.1007/s00284-020-02032-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02032-0