Abstract
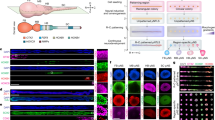
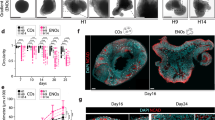
The study of brain development in humans is limited by the lack of tissue samples and suitable in vitro models. Here, we model early human neural tube development using human embryonic stem cells cultured in a microfluidic device. The approach, named microfluidic-controlled stem cell regionalization (MiSTR), exposes pluripotent stem cells to signaling gradients that mimic developmental patterning. Using a WNT-activating gradient, we generated a neural tissue exhibiting progressive caudalization from forebrain to midbrain to hindbrain, including formation of isthmic organizer characteristics. Single-cell transcriptomics revealed that rostro-caudal organization was already established at 24 h of differentiation, and that the first markers of a neural-specific transcription program emerged in the rostral cells at 48 h. The transcriptomic hallmarks of rostro-caudal organization recapitulated gene expression patterns of the early rostro-caudal neural plate in mouse embryos. Thus, MiSTR will facilitate research on the factors and processes underlying rostro-caudal neural tube patterning.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
scRNAseq data is deposited in the Gene Expression Omnibus with accession number GSE135399. scRNAseq code used for analysis is supplied as Supplementary Code in html and .rmd files and can also be found on GitHub (https://github.com/kirkeby-lab).
Change history
11 June 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Nordstrom, U., Jessell, T. M. & Edlund, T. Progressive induction of caudal neural character by graded Wnt signaling. Nat. Neurosci. 5, 525–532 (2002).
Kiecker, C. & Lumsden, A. Compartments and their boundaries in vertebrate brain development. Nat. Rev. Neurosci. 6, 553–564 (2005).
Kiecker, C. & Lumsden, A. The role of organizers in patterning the nervous system. Annu Rev. Neurosci. 35, 347–367 (2012).
Ribes, V. & Briscoe, J. Establishing and interpreting graded Sonic Hedgehog signaling during vertebrate neural tube patterning: the role of negative feedback. Cold Spring Harb. Perspect. Biol. 1, a002014 (2009).
Meinhardt, A. et al. 3D reconstitution of the patterned neural tube from embryonic stem cells. Stem Cell Rep. 3, 987–999 (2014).
Demers, C. J. et al. Development-on-chip: in vitro neural tube patterning with a microfluidic device. Development 143, 1884–1892 (2016).
Bagley, J. A., Reumann, D., Bian, S., Levi-Strauss, J. & Knoblich, J. A. Fused cerebral organoids model interactions between brain regions. Nat. Methods 14, 743–751 (2017).
Birey, F. et al. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59 (2017).
Cederquist, G. Y. et al. Specification of positional identity in forebrain organoids. Nat. Biotechnol. 37, 436–444 (2019).
Kirkeby, A. et al. Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell Rep. 1, 703–714 (2012).
Jeon, N. L. et al. Generation of solution and surface gradients using microfluidic systems. Langmuir 16, 8311–8316 (2000).
Chambers, S. M. et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280 (2009).
Dupe, V. et al. In vivo functional analysis of the Hoxa-1 3’ retinoic acid response element (3’RARE). Development 124, 399–410 (1997).
Strate, I., Min, T. H., Iliev, D. & Pera, E. M. Retinol dehydrogenase 10 is a feedback regulator of retinoic acid signalling during axis formation and patterning of the central nervous system. Development 136, 461–472 (2009).
Yang, L. et al. Analysis of FGF-dependent and FGF-independent pathways in otic placode induction. PLoS ONE 8, e55011 (2013).
La Manno, G. et al. Molecular diversity of midbrain development in mouse, human, and stem cells. Cell 167, 566–580 (2016).
Pijuan-Sala, B. et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495 (2019).
Quadrato, G. et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545, 48–53 (2017).
Kanton, S. et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 574, 418–422 (2019).
Stoeckius, M. et al. Cell hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19, 224 (2018).
Brafman, D. & Willert, K. Wnt/beta-catenin signaling during early vertebrate neural development. Dev. Neurobiol. 77, 1239–1259 (2017).
Metzis, V. et al. Nervous system regionalization entails axial allocation before neural differentiation. Cell 175, 1105–1118 (2018).
Abu-Abed, S. et al. The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev. 15, 226–240 (2001).
Andoniadou, C. L. et al. HESX1- and TCF3-mediated repression of Wnt/beta-catenin targets is required for normal development of the anterior forebrain. Development 138, 4931–4942 (2011).
Peng, G. & Westerfield, M. Lhx5 promotes forebrain development and activates transcription of secreted Wnt antagonists. Development 133, 3191–3200 (2006).
Filipe, M., Goncalves, L., Bento, M., Silva, A. C. & Belo, J. A. Comparative expression of mouse and chicken Shisa homologues during early development. Dev. Dyn. 235, 2567–2573 (2006).
Xia, Y. & Whitesides, G. M. Soft Lithography. Angew. Chem. Int. Ed. 37, 550–575 (1998).
Satyanarayana, S., Karnik, R. N. & Majumdar, A. Stamp-and-stick room-temperature bonding technique for microdevices. J. Microelectromechanical Syst. 14, 392–399 (2005).
Edward, J. T. Molecular volumes and the Stokes–Einstein equation. J. Chem. Educ. 47, 261 (1970).
Maury, Y. et al. Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat. Biotechnol. 33, 89–96 (2015).
Nestorowa, S. et al. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood 128, e20–31 (2016).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902(2019).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Haghverdi, L., Lun, A. T. L., Morgan, M. D. & Marioni, J. C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36, 421–427 (2018).
Kempf, H. et al. Bulk cell density and Wnt/TGFbeta signalling regulate mesendodermal patterning of human pluripotent stem cells. Nat. Commun. 7, 13602 (2016).
Funa, N. S. et al. beta-Catenin regulates primitive streak induction through collaborative interactions with SMAD2/SMAD3 and OCT4. Cell Stem Cell 16, 639–652 (2015).
Acknowledgements
This study was supported by the Novo Nordisk Foundation (grant no. NNF18OC0030286 to A.K.), The Lundbeck Foundation (grant no. R190-2014-3904 to T.H.P.) and the following grants to A.K.: Innovation Fund Denmark (no. BrainStem 4108-00008 A), the Strong Research Environment at Lund University Multipark, the Swedish Research Council (no. 70862601/Bagadilico), The Crafoord Foundation, The Segerfalk Foundation, The Tore Nilsson Foundation, The Sven-Olof Janson Foundation and the Swedish Fund for Research Without Animal Experiments. The research leading to these results has received funding from the New York Stem Cell Foundation (M.P.), the European Research Council under the ERC Grant Agreement no. 30971 (M.P.), the Swedish Research Council (grant agreement no. 521-2012-5624, M.P.). The Novo Nordisk Foundation Center for Stem Cell Biology (DanStem) and the Novo Nordisk Foundation Center for Basic Metabolic Research (CBMR) are supported by Novo Nordisk Foundation grants (nos. NNF17CC0027852 and NNF18CC0034900, respectively). M.P. is a New York Stem Cell Foundation Robertson Investigator. We thank S. da Rocha Baez, I. Nilsson, M. Madrona, M. Heide Ankjær, H.K. Lilja-Fischer (CBMR Single-cell Omics Platform), H. Neil (DanStem Genomics Platform), J. Bulkescher (DanStem Imaging Platform) and A. Meligkova (DanStem Stem Cell Culture Platform) for excellent technical and bioinformatics assistance and for use of instruments.
Author information
Authors and Affiliations
Contributions
M.I., T.L., P.R., T.H.P., M.P., G.S.R. and A.K. designed the study. M.I., P.R., G.S.R., P.A.K., D.M.R., G.B., K.L.E., O.K.M. and J.L. performed experiments. M.I., T.L., P.R. and A.K. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8, Tables 3–5 and Notes.
Supplementary Table 1
Marker genes for cell clusters obtained from single-cell RNAseq analysis
Supplementary Table 2
Supplementary statistical summary of analysis
Rights and permissions
About this article
Cite this article
Rifes, P., Isaksson, M., Rathore, G.S. et al. Modeling neural tube development by differentiation of human embryonic stem cells in a microfluidic WNT gradient. Nat Biotechnol 38, 1265–1273 (2020). https://doi.org/10.1038/s41587-020-0525-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-020-0525-0
This article is cited by
-
Genetics of human brain development
Nature Reviews Genetics (2024)
-
A patterned human neural tube model using microfluidic gradients
Nature (2024)
-
Single-cell Profiling of Reprogrammed Human Neural Stem Cells Unveils High Similarity to Neural Progenitors in the Developing Central Nervous System
Stem Cell Reviews and Reports (2024)
-
Humanized brain organoids-on-chip integrated with sensors for screening neuronal activity and neurotoxicity
Microchimica Acta (2024)
-
Identifying secreted biomarkers of dopaminergic ventral midbrain progenitor cells
Stem Cell Research & Therapy (2023)