Abstract
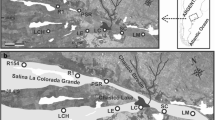
Understanding the processes and patterns of local adaptation and migration involves an exhaustive knowledge of how landscape features and population distances shape the genetic variation at the geographical level. Ctenomys australis is an endangered subterranean rodent characterized by having a restricted geographic range immerse in a highly fragmented sand dune landscape in the Southeast of Buenos Aires province, Argentina. We use 13 microsatellite loci in a total of 194 individuals from 13 sampling sites to assess the dispersal patterns and population structure in the complete geographic range of this endemic species. Our analyses show that populations are highly structured with low rates of gene flow among them. Genetic differentiation among sampling sites was consistent with an isolation by distance pattern, however, an important fraction of the population differentiation was explained by natural barriers such as rivers and streams. Although the individuals were sampled at locations distanced from each other, we also use some landscape genetics approaches to evaluate the effects of landscape configuration on the genetic connectivity among populations. These analyses showed that the sand dune habitat availability (the most suitable habitat for the occupation of the species), was one of the main factors that explained the differentiation patterns of the different sampling sites located on both sides of the Quequén Salado River. Finally, habitat availability was directly associated with the width of the sand dune landscape in the Southeast of Buenos Aires province, finding the greatest genetic differentiation among the populations of the Northeast, where this landscape is narrower.





Similar content being viewed by others
References
Apfelbaum LI, Massarini AI, Daleffe LE, Reig OA (1991) Genetic variability in the subterranean rodents Ctenomys australis and Ctenomys porteusi (Rodentia: Octodontidae). Biochem Syst Ecol 19:467–476
Apodaca JJ, Rissler LJ, Godwin JC (2012) Population structure and gene flow in a heavily disturbed habitat: implications for the management of the imperilled Red Hills salamander (Phaeognathus hubrichti). Conserv Genet 13:913–923
Balding DJ, Nichols RA (1995) A method for quantifying differentiation between populations at multi-allelic loci and its implications for investigating identity and paternity. Genetica 96:3–12
Beerli P (2006) Comparison of Bayesian and maximum likelihood inference of population genetic parameters. Bioinformatics 22:341–345
Beerli P (2008) MIGRATE documentation (version 3.0). Technical Report
Beerli P, Felsenstein J (2001) Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc Natl Acad Sci USA 98:4563–4568
Bergek S, Björklund M (2007) Cryptic barriers to dispersal within a lake allow genetic differentiation within a population of Eurasian perch (Perca fluviatilis). Evolution 61:2035–2041
Biello R, Brunelli A, Sozio G, Havenstein K, Mortelliti A, Ketmaier V, Bertorelle G (2018) Genetic structure in the wood mouse and the bank vole: contrasting patterns in a human-modified and highly fragmented landscape. bioRxiv. https://doi.org/10.1101/464057
Björklund M, Ruiz I, Senar JC (2010) Genetic differentiation in the urban habitat: the great tits (Parus major) of the parks of Barcelona city. Biol J Linn Soc 99:9–19
Bruvo R, Michiels NK, D’Souza TG, Schulenburg H (2004) A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Mol Ecol 13:2101–2106
Busch C, Antinuchi CD, del Valle JC, Kittlein MJ, Malizia AI, Vassallo AI, Zenuto RR (2000) Population ecology of subterranean rodents. In: Lacey EA, Patton JL, Cameron GN (eds) Life underground: the biology of subterranean rodents. University of Chicago Press, Chicago, pp 183–226
Celsi CE, Monserrat AL (2008) La vegetación dunícola en el frente costero de la Pampa Austral (Partido de Coronel Dorrego, Buenos Aires). Multequina 17:73–92
Chiucchi JE, Gibbs HL (2010) Similarity of contemporary and historical gene flow among highly fragmented populations of an endangered rattlesnake. Mol Ecol 19:5345–5358
Ciofi C, Beaumont MA, Swingland IR, Bruford MW (1999) Genetic divergence and units for conservation in the Komodo dragon Varanus komodoensis. Proc R Soc Lond B 266:2269–2274
Cornuet J, Piry S, Luikart G, Estoup A, Solignac M (1999) New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics 153:1989–2000
Crispo E, Moore JS, Lee-Yaw JA, Gray SM, Haller BC (2011) Broken barriers: human-induced changes to gene flow and introgression in animals. BioEssays 33:508–518
Crooks KR, Sanjayan M (2006) Connectivity conservation. Cambridge University Press, Cambridge
Cutrera AP, Mora MS (2017) Selection on MHC in a Context of Historical Demographic Change in 2 Closely Distributed Species of Tuco-tucos (Ctenomys australis and C. talarum). J Hered 108:628–639
Cutrera AP, Mora MS, Antenucci CD, Vassallo AI (2010) Intra and interspecific variation in home-range size in sympatric tuco-tucos, Ctenomys australis and C. talarum. J Mammal 91(6):1425–1434
Dionne M, Caron F, Dodson JJ, Bernatchez L (2008) Landscape genetics and hierarchical genetic structure in Atlantic salmon: the interaction of gene flow and local adaptation. Mol Ecol 17:2382–2396
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals structure: a simulation study using the software. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2005) ARLEQUIN ver. 3.0: an integrated software packaged for population genetics data analysis. Evol Bioinform 1:47–50
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Faubet P, Waples RS, Gaggiotti OE (2007) Evaluating the performance of a multilocus Bayesian method for the estimation of migration rates. Mol Ecol 16:1149–1166
Fernández-Stolz GP, Stolz JFB, de Freitas TRO (2007) Bottlenecks and dispersal in the tuco-tuco das dunas, Ctenomys flamarioni (Rodentia: Ctenomyidae): in Southern Brazil. J Mammal 88(4):935–945
Foll M, Gaggiotti OE (2006) Identifying the environmental factors that determine the genetic structure of populations. Genetics 174:875–891
Galiano D, Bernardo-Silva J, de Freitas TRO (2014) Genetic pool information reflects highly suitable areas: the case of two parapatric endangered species of tuco-tucos (Rodentia: Ctenomyidae). PLoS ONE 9:e97301
Gonçalves GL, Freitas TRO (2009) Intraspecific variation and genetic differentiation of the collared tuco-tuco (Ctenomys torquatus) in Souther Brazil. J Mammal 90(4):1020–1031
Guo S, Thompson E (1992) Performing the exact test of Hardy-Weinberg proportion for multiples alleles. Biometrics 48:361–372
Hanski I, Gaggiotti OE (2004) Ecology, genetics, and evolution of metapopulations. Elsevier Academic Press, San Diego
Isla FI, Cortizo LC, Turno OH (2001) Dinámica y Evolución de las Barreras Medanosas, Provincia de Buenos Aires, Argentina. Rev Bras Geomorfol 2:73–83
Isla FI, Bértola G, Merlotto A, Ferrante A, Cortizo L (2009) Requerimientos y disponibilidad de arenas para la defensa de las playas de Necochea y Loberia. Rev Asoc Geol Argentina 65:446–456
Kalinowski ST (2011) The computer program STRUCTURE does not reliably identify the main genetic clusters within species: simulations and implications for human population structure. Heredity 106:625–632
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1100
Kamvar ZN, Tabima JF, Grünwald NJ (2014) Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281. https://doi.org/10.7717/peerj.281
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Kierepka EM, Anderson SJ, Swihart RK, Rhodes OE Jr (2016) Evaluating the influence of life-history characteristics on genetic structure: a comparison of small mammals inhabiting complex agricultural landscapes. Ecol Evol 6:6376–6396
Kittlein MJ, Vassallo AI, Mora MS, de Durana F, Ricciardulli MG, Tizón FR (2004) Dunas del Sureste Bonaerense. In: Bilenca D, Miñarro F (eds) Identificación de Áreas Valiosas de Pastizal en las Pampas y Campos de Argentina. Uruguay y Sur de Brasil, Fundación Vida Silvestre Argentina, Buenos Aires, pp 76–77
Lacey EA (2000) Spatial and social systems of subterranean rodents. In: Lacey EA, Patton JL, Cameron GN (eds) Life Underground: the biology of subterranean rodents. University of Chicago Press, Chicago and London, pp 257–293
Lacey EA (2001) Microsatellite variation in solitary and social tuco-tucos: molecular properties and population dynamics. Heredity 86:628–637
Lacey EA, Maldonado JE, Clabaugh JP, Matocq MD (1999) Interspecific variation in microsatellites isolated from tuco-tucos (Rodentia: Ctenomyidae). Mol Ecol 8:1753–1768
Lopes CM (2011) História evolutiva de Ctenomys minutus e Ctenomys lami (Rodentia, Ctenomyidae) na planície costeira do Sul do Brasil. [PhD thesis]. [Porto Alegre (Brazil)]: Universidade Federal do Rio Grande do Sul.
Manier MK, Arnold SJ (2005) Population genetic analysis identifies source-sink dynamics for two sympatric garter snake species (Thamnophis elegans and Thamnophis sirtalis). Mol Ecol 14:3965–3976
Mantel N (1967) The detection of disease clustering and a generalized regression approaches. Cancer Res 27:209–220
Mapelli FJ, Kittlein MJ (2009) Influence of patch and landscape characteristics on the distribution of the subterranean rodent Ctenomys porteousi. Landsc Ecol 24(6):726–733
Mapelli FJ, Mora MS, Mirol PM, Kittlein MJ (2012) Population structure and landscape genetics in the endangered subterranean rodent Ctenomys porteousi. Conserv Genet 13:165–181
Marcomini SC, López RA (2016) Geología de la Costa Marina Bonaerense. In: Athor J, Celsi CE (eds) La Costa Atlántica de Buenos Aires: Naturaleza y Patrimonio Cultural. Fundación de Historia Natural Félix de Azara, Buenos Aires, pp 20–41
McGarigal K, Cushman SA, Neel MC, Ene E (2002) FRAGSTATS: Spatial pattern analysis program for categorical maps. University of Massachusetts, Amherst
McGarigal K, Cushman SA, Ene E (2012) FRAGSTATS v4: spatial pattern analysis program for categorical and continuous maps. University of Massachusetts, Amherst
Meirmans PG (2015) Seven common mistakes in population genetics and how to avoid them. Mol Ecol 24:3223–3231
Mora MS, Mapelli FJ (2010) Conservación en médanos: Fragmentación del hábitat y dinámica poblacional del tuco–tuco de las dunas. In: Isla FI, Lasta CA (eds) Manual de manejo de barreras medanosas de la Provincia de Buenos Aires. Universidad de Mar del Plata, Mar del Plata, pp 161–181
Mora MS, Lessa EP, Kittlein MJ, Vassallo AI (2006) Phylogeography of the subterranean rodent Ctenomys australis in sand-dune habitats: evidence of population expansion. J Mammal 87:1192–1203
Mora MS, Lessa EP, Cutrera AP, Kittlein MJ, Vassallo AI (2007) Phylogeographical structure in the subterranean tuco-tuco Ctenomys talarum (Rodentia: Ctenomyidae): contrasting the demographic consequences of regional and habitat-specific histories. Mol Ecol 16:3453–3465
Mora MS, Mapelli FJ, Gaggiotti OE, Kittlein MJ, Lessa EP (2010) Dispersal and population structure at different spatial scales in the subterranean rodent Ctenomys australis. BMC Genet 11:1–14
Mora MS, Cutrera AP, Lessa EP, Vassallo AI, D’Anatro A, Mapelli FJ (2013) Phylogeography and population genetic structure of the Talas tuco-tuco (Ctenomys talarum): integrating demographic and habitat histories. J Mammal 94(2):459–476
Mora MS, Mapelli FJ, López A, Gómez Fernández MJ, Mirol PM, Kittlein MJ (2016) Population genetic structure and historical dispersal patterns in the subterranean rodent Ctenomys “chasiquensis” from the southeastern Pampas region. Argentina Mammal Biol 81(3):314–325
Mora MS, Mapelli FJ, López A, Gómez Fernández MG, Mirol PM, Kittlein MJ (2017) Landscape genetics in the subterranean rodent Ctenomys “chasiquensis” associated with highly disturbed habitats from the southeastern Pampas region, Argentina. Genetica 145:575–591
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Paetkau D, Calvert W, Stirling I, Strobeck C (1995) Microsatellite analysis of population structure of Canadian polar bears. Mol Ecol 4:347–354
Paetkau D, Slade R, Burden M, Estoup A (2004) Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Mol Ecol 13:55–65
Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecol Model 190:231–259
Pritchard JK, Wen W (2003) Documentation for STRUCTURE software: Version 2. https://www.pritch.bsd.uchicago.edu
Pritchard J, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rambaut A, Suchard MA, Xie W, Drummond AJ (2014) Version v1.6.0. https://beast.bio.ed.ac.uk/Tracer
Rannala B, Mountain JL (1997) Detecting immigration by using Multilocus genotypes. Proc Natl Acad Sci USA 94:9197–9201
Rico C, Cuesta JA, Drake P, Macpherson E, Bernatchez L, Marie AD (2017) Null alleles are ubiquitous at microsatellite loci in the Wedge Clam (Donax trunculus). PeerJ 5:e3188. https://doi.org/10.7717/peerj.3188
Roratto PA, Bartholomei-Santos ML, de Freitas TRO (2011) Tetranucleotide microsatellite markers in Ctenomys torquatus (Rodentia). Conserv Genet Resour 3:725–727
Roratto PA, Fernandes FA, de Freitas TRO (2014) Phylogeography of the subterranean rodent Ctenomys torquatus: an evaluation of the riverine barrier hypothesis. J Biogeogr 42:694–705
Sato JJ, Kawakami T, Tasaka Y, Tamenishi M, Yamaguchi Y (2014) A few decades of habitat fragmentation has reduced population genetic diversity: a case study of landscape genetics of the large Japanese field mouse, Apodemus speciosus. Mammal Study 39:1–10
Sikes RS, Animal Care and Use Committee of the American Society of Mammalogists (2016) Guidelines of the American Society of Mammalogists for the use of wild mammals in research and education. J Mammal 97:663–688
Slatkin M (1993) Isolation by distance in equilibrium and nonequilibrium populations. Evolution 47:264–279
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462
Spiegelhalter DJ, Best NG, Carlin BR, van der Linde A (2002) Bayesian measures of model complexity and fit. J R Stat Soc B 64:583–616
Steinberg EK, Patton JL (2000) Genetic structure and the geographic of the speciation in subterranean rodents: opportunities and constraints for evolutionary diversification. In: Lacey EA, Patton JL, Cameron GN (eds) Life underground: the biology of subterranean rodents. University of Chicago Press, Chicago, pp 183–226
Sutherland GD, Harestad AS, Price K, Lertzman KP (2000) Scaling of natal dispersal distances in terrestrial birds and mammals. Conserv Ecol 4:16
Tomasco IT, Lessa EL (2007) Phylogeography of the Tuco-tuco Ctenomys pearsoni: mtDNA variation and its Implication for Chromosomal Differentiation. In: Kelt DA, Lessa EP, Salazar-Bravo J, Patton JL (eds) The quintessential naturalist: honoring the life and legacy of Oliver P. University of California Press, Pearson, Berkeley, Los Angeles and London, pp 859–882
Turno Orellano HA, Isla FI, Juárez VI (2003) Implementación de un SIG en la evaluación de la aptitud para prácticas forestales en el litoral bonaerense. Boletim Paranaense de Geociências 53:27–34
Turno Orellano HA, Isla FI (2004) Developing sinks for CO2 through forestation of temperate coastal barriers: an environmental business. Reg Environ Change 4:70–76
Vassallo AI (1998) Functional morphology, comparative behaviour, and adaptation in two sympatric subterranean rodents of genus Ctenomys (Caviomorpha: Octodontidae). J Zool 244:415–427
Visser JH, Bennett NC, van Vuuren BJ (2018) Spatial genetic diversity in the Cape mole-rat, Georychus capensis: Extreme isolation of populations in a subterranean environment. PLoS ONE 13:e0194165
Waples RS, Do C (2010) Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol Appl 3:244–262
Waples R, Gaggiotti OE (2006) What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol Ecol 15:1419–1439
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191
Wlasiuk G, Garza JC, Lessa EP (2003) Genetic and geographic differentiation in the Rio Negro tuco-tuco (Ctenomys rionegrensis): inferring the roles of migration and drift from multiple genetic markers. Evolution 57:913–926
Zenuto RR, Busch C (1995) Influence of the subterranean rodent Ctenomys australis (tuco-tuco) in a sand-dune grassland. Z Säugetierkunde 60:277–285
Zenuto RR, Busch C (1998) Population biology of the subterranean rodent Ctenomys australis (tuco-tuco) in a coastal dune-field in Argentina. Z Säugetierkunde 63:357–367
Acknowledgements
We are grateful to all the members of the Museo Argentino de Ciencias Naturales “Bernardino Rivadavia” for their invaluable support and advice. Financial support was provided by Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, PIP-11220150100066CO), UNMdP (Project EXA903/18) and FONCYT (PICT-201-0427). To these persons and institutions, we express our deep gratitude.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Austrich, A., Mora, M.S., Mapelli, F.J. et al. Influences of landscape characteristics and historical barriers on the population genetic structure in the endangered sand-dune subterranean rodent Ctenomys australis. Genetica 148, 149–164 (2020). https://doi.org/10.1007/s10709-020-00096-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-020-00096-1