Abstract
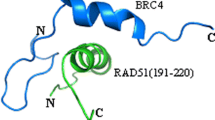
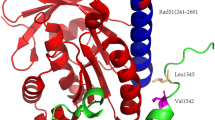
BRCA2 is an important tumor suppressor gene that plays a critical role in preserving the stability of cellular genetic information, participating in DNA repair by engaging in binding interactions with RAD51 proteins. However, the lack of structural data on BRCA2 and RAD51 makes the study of their interaction mechanism still a great challenge. We characterize the structure of the BRC8–RAD51 complex using ZDOCK protein docking software and identify the potential non-conserved active site of BRC8 via virtual alanine scanning, utilizing the obtained results to synthesize BRC8, its six analogous peptides (BRC8-1 to BRC8-6), and critical peptide fragment of RAD51 (RAD51(231–260)) by Fmoc solid-phase synthesis. The analogous peptides are found to exhibit a secondary structure significantly different from that of BRC8 by circular dichroism spectroscopy, which indicates that mutation sites determined by computer-aided simulation correspond to key amino acid residues substantially affecting polypeptide structure. On the other hand, the secondary structure of RAD51(231–260) was also considerably influenced by its interaction with BRC8 and analogs, e.g., the fraction of the α-helical structure in RAD51(231–260) increased to 23.6, 15.1, and 13.5% upon interaction with BRC8-1, BRC8-3, and BRC8-6, respectively. The results show that the properties of C-terminal amino acid residues significantly influence peptide–peptide interactions, in agreement with the results of virtual alanine scanning. Therefore, computer-aided simulation was confirmed to be a technique that is useful for narrowing down the range of sites responsible for interactions between peptides or proteins, and provides new inspirations for the design of peptides with strong interactions.




Similar content being viewed by others
References
Ahmed A, Shamsi A, Khan MS, Husain FM, Bano B (2018) Methylglyoxal induced glycation and aggregation of human serum albumin: biochemical and biophysical approach. Int J Biol Macromol 113:269–276
Carreira A (2009) The BRC repeats of BRCA2 modulate the DNA-binding specificity of RAD51. Cell 136:1032–1043
Ciemny M, Kurcinski M, Kamel K, Kolinski A, Alam N, Schueler-Furman O, Kmiecik S (2018) Protein-peptide docking: opportunities and challenges. Drug Discov Today 23:1530–1537
Dagan T, Talmor Y, Graur D (2002) Ratios of radical to conservative amino acid replacement are affected by mutational and compositional factors and may not be indicative of positive darwinian selection. Mol Biol Evol 19:1022–1025
Cole DJ, Rajendra E, Roberts-Thomson M, Hardwick B, McKenzie GJ, Payne MC, Venkitaraman AR, Skylaris C (2011) Interrogation of the protein-protein interactions between human BRCA2 BRC repeats and RAD51 reveals atomistic determinants of affinity. PloS Comput Biol 7(7):e1002096
Esashi F, Galkin VE, Yu X, Egelman EH, West SC (2007) Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol 14:468–474
Feng KX, Lu ZZ, Pang C, Yun WY (2018) An efficient computational method of a moment-independent importance measure using quantile regression. Mech Syst Signal Pr 109:235–246
Gudiukaite R, Gegeckas A, Sadauskas M, Citavicius D (2016) Detection of Asp371, Phe375, and Tyr376 influence on GD-95-10 lipase using alanine scanning mutagenesis. Appl Biochem Biotech 178:654–669
Haider MK, Bertrand HO, Hubbard RE (2011) Predicting fragment binding poses using a combined MCSS MM-GBSA approach. J Chem Inf Model 51:1092–1105
Harper JW, Elledge SJ (2007) The DNA damage response: ten years after. Mol Cell 28:739–744
Hattori H, Janky R, Nietfeld W, Aerts S, Madan BM, Venkitaraman AR (2014) p53 shapes genome-wide and cell type-specific changes in microRNA expression during the human DNA damage response. Cell Cycle 13:2572–2586
Hengel SR, Spies MA, Spies M (2017) Small-molecule inhibitors targeting DNA repair and DNA repair deficiency in research and cancer therapy. Cell Chem Biol 24(9):1101–1119
Jensen RB, Carreira A, Kowalczykowski SC (2010) Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467:678–683
Junaid M, Muhseen ZT, Ullah A, Wadood A, Liu J, Zhang H (2014) Molecualr modeling and molecular dynamics simulation study of the human Rab9 and RhoBTB3 C-terminus complex. Bioinformation 10:757–763
Kan C, Zhang JR (2015) BRCA1 mutation: a predictive marker for radiation therapy? Int J Radiat Oncol 93:281–293
Karhu L, Turku A, Xhaard H (2015) Modeling of the OX1R-orexin-A complex suggests two alternative binding modes. BMC Struct Biol 158:9
Kelly SM, Price NC (2000) The use of circular dichroism in the investigation of protein structure and function. Curr Protein Pept Sci 1:349–384
Li HL, Ma Y, Ma Y, Li Y, Chen XB, Dong WL, Wang RL (2017) The design of novel inhibitors for treating cancer by targeting CDC25B through disruption of CDC25B-CDK2/Cyclin a interaction using computational approaches. Oncotarget 8:33225–33240
Lighezan L, Georgieva R, Neagu A (2016) The secondary structure and the thermal unfolding parameters of the S-layer protein from Lactobacillus salivarius. Eur Biophys J Biophy 45:491–509
Lippow SM, Tidor B (2007) Progress in computational protein design. Curr Opin Biotech 18:305–311
Martinez JS, Von NC, Kim T, Ehlen A, Mazin AV, Kowalczykowski SC, Carreira A (2016) BRCA2 regulates DMC1-mediated recombination through the BRC repeats. P Natl Acad Sci USA 113:515–520
Moreira IS, Fernandes PA, Ramos MJ (2006) Unraveling the importance of protein-protein interaction: application of a computational alanine-scanning mutagenesis to the study of the IgG1 strptococcal protein G (C2 fragment) complex. J Phys Chem 110:10962–10969
Mortins SA, Perez MAS, Moreira IS, Sousa SF, Ramos MJ, Fernandes PA (2013) Computational alanine scanning mutagenesis: MM-PBSA vs TI. J Chem Theory Comput 9:1311–1319
Nathanson KN, Wooster R, Weber BL (2001) Breast cancer genetics, what we know and what we need. Nat Med 7:552–556
Neco AHB, Pinto-Junior VR, Araripe DA, Santiago MQ, Osterne VJS, Lossio CF, Nobre CAS, Oliveira MV, Silva MTL, Martins MGQ, Cajazeiras JB, Marques GFO, Costa DR, Nascimento KS, Assreuy AMS, Cavada BS (2018) Structural analysis, molecualr docking and molecular dynamics of an edematogenic lectin from Centrolobium microchaete seeds. Int J Biol Macromol 117:124–133
Nomme J, Renodon-Corniѐre A, Asanomi Y, Sakaguchi K, Stasiak AZ, Stasiak A, Norden B, Tran V, Takahashi M (2010) Design of potent inhibitors of human RAD51 recombinase based on BRC motifs of BRCA2 protein: modeling and experimental validation of chimera peptide. J Med Chem 53:5782–5791
Otvos L Jr, Wade JD (2014) Current challenges in peptide-based drug discovery. Front Chem 2:62
Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR (2002) Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature 420:287–293
Poulos RC, Olivier J, Wong JWH (2017) The interaction between cytosine methylation and processes of DNA replication and repair shape the mutation lanscape of cancer genomes. Nucleic Acids Res 45:7786–7795
Powell SN, Kachnic LA (2003) Roles of BRCA1 and BRCA2 in homologous recombination, DNA replication fidelity and the cellular response to ionizing Radiation. Oncogene 37:5784–5791
Shen MY, Sali A (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci 15:2507–2524
Sobocinska M, Gieldon A, Fichna J, Kamysz E (2018) Alanine scan of sialorphin and its hybrids with opiorphin: synthesis, molecular modelling and effect on enkephalins degradation. Amino Acids 50:1083–1088
Subramanyam S, Jones WT, Spies M, Spies MA (2013) Contributions of the RAD51 N-terminal domain to BRCA2–RAD51 interaction. Nucleic Acids Res 41(19):9020–9032
Thorslund T (2010) The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol 17:1263–1265
Waterworth A (2003) New insights into the biological function of BRCA2 from its structural interactions. Breast Cancer Res 5:107–108
Wulan DR, Utomo EP, Mahdi C (2014) Molecular modeling of Ruellia tuberosal L compounds as a-amylase inhibitor: an in silico comparation between human and rat enzyme model. Bioinformation 10:209–215
Zhao DX, Lu K, Liu GB, Ma L, Zhu HJ, He J (2018) Design and synthesis of BRC analogous peptides and their interactions with a key p53 peptide. FEBS Lett 592(20):3438–3445
Funding
This work was supported by the National Natural Science Foundation of China (grant number 21572046) and Key Scientific Research Project of Henan Higher Education Institutions (grant number 20B150006).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human and animal participants
This article does not contain any studies with human participants performed by any of authors.
Additional information
Handling Editor: V. Soloshonok.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, D., Lu, K., Liu, G. et al. Design of BRC analogous peptides based on the complex BRC8–RAD51 and the preliminary study on the peptide structures. Amino Acids 52, 831–839 (2020). https://doi.org/10.1007/s00726-020-02856-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-020-02856-x