Abstract
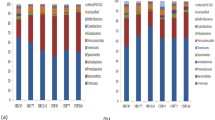
The objective of this study was to evaluate the effect of undernutrition on colonic microbiota and fermentation in pregnant ewes. Sixteen ewes bearing multiple fetuses for 115 days in the control (CON) and severe feed restriction (SFR) groups were fed 100% and 30% level of ad libitum feed intake, respectively. After 15-day treatment, all ewes were sacrificed to collect colonic digesta samples to extract DNA for 16S rRNA sequencing and to detect fermentation parameters. Our data showed that SFR increased (P < 0.05) the levels of colonic propionate, isobutyrate, butyrate, isovalerate, and valerate, and slightly decreased (P < 0.1) colonic pH. The mole proportions of isobutyrate, butyrate, and isovalerate were increased (P < 0.05) upon SFR while that of acetate was decreased (P < 0.05). Hematoxylin-eosin staining sections exhibited the disorderly, irregular, and loose arrangement and part sloughing of colonic epithelial cells. Furthermore, SFR decreased (P < 0.05) the diversity of colonic microbiota and changed the microbial communities. At the genus level, SFR increased (P < 0.05) the abundance of unclassified Peptococcaceae and decreased (P < 0.05) the abundances of Ruminococcus, unclassified Ruminococcaceae, and unclassified VadinBB60. Additionally, the abundances of Ruminococcus and unclassified Ruminococcaceae were positively correlated (P < 0.05) with the acetate proportion while the abundance of unclassified Peptococcaceae was negatively correlated (P < 0.05) with the percentages of isobutyrate, butyrate, and isovalerate. In summary, SFR diminished the diversity of bacteria, affected the composition of bacterial communities, and finally changed the colonic fermentation pattern and epithelial histomorphology in pregnant ewes.
Key Points
• Undernutrition changed colonic bacterial diversity and composition in pregnant ewes.
• Microbial alteration affected colonic fermentation pattern and parameters.
• Alteration of colonic microbiota and fermentation damaged epithelium histomorphology.

Graphical abstract







Similar content being viewed by others
References
Azad E, Derakhshani H, Forster R, Gruninger R, Acharya S, McAllister T, Khafipour E (2019) Characterization of the rumen and fecal microbiome in bloated and non-bloated cattle grazing alfalfa pastures and subjected to bloat prevention strategies. Sci Rep 9(1):4272
Baxter NT, Schmidt AW, Venkataraman A, Kim KS, Waldron C, Schmidt TM (2019) Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. MBio 10(1):e02566–e02518
Cal-Pereyra L, Acosta-Dibarrat J, Benech A, Da Silva S, Martín A, González-Montaña JR (2012) Toxemia de la gestación en ovejas: revisión. Rev Mex Cienc Pecu 3(2):247–264
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R (2009) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26(2):266–267
Carvalho R, Vaz A, Pereira FL, Dorella F, Aguiar E, Chatel J-M, Bermudez L, Langella P, Fernandes G, Figueiredo H (2018) Gut microbiome modulation during treatment of mucositis with the dairy bacterium Lactococcus lactis and recombinant strain secreting human antimicrobial PAP. Sci Rep 8(1):15072
Christopherson MR, Dawson JA, Stevenson DM, Cunningham AC, Bramhacharya S, Weimer PJ, Kendziorski C, Suen G (2014) Unique aspects of fiber degradation by the ruminal ethanologen Ruminococcus albus 7 revealed by physiological and transcriptomic analysis. BMC Genomics 15(1):1066
Crost E, Le Gall DG, Laverde-Gomez J, Mukhopadhya I, Flint H, Juge N (2018) Mechanistic insights into the cross-feeding of Ruminococcus gnavus and Ruminococcus bromii on host and dietary carbohydrates. Front Microbiol 9:2558
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10(10):996–998
Forbes J (1969) The effect of pregnancy and fatness on the volume of rumen contents in the ewe. J Agr Sci 72(1):119–121
Forbes J (1970) The voluntary food intake of pregnant and lactating ruminants: a review. Br Vet J 126:1–11
Forbes JM, Dquo RJ, Boaz T (1967) Silage as a feed for pregnant ewes. Anim Sci 9(3):399–408
Genton L, Cani PD, Schrenzel J (2015) Alterations of gut barrier and gut microbiota in food restriction, food deprivation and protein-energy wasting. Clin Nutr 34(3):341–349
Gordon J, Tribe D (1951) The self-selection of diet by pregnant ewes. J Agr Sci 41(1–2):187–190
Guinane CM, Cotter PD (2013) Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Ther Adv Gastroenter 6(4):295–308
Hadjipieris G, Holmes W (1966) Studies on feed intake and feed utilization by sheep I. Voluntary feed intake of dry, pregnant and lactating ewes. J Agr Sci 66(2):217–223
Hu F, Xue Y, Guo C, Liu J, Mao S (2018) The response of ruminal fermentation, epithelium-associated microbiota, and epithelial barrier function to severe feed restriction in pregnant ewes. J Anim Sci 96(10):4293–4305
Huws SA, Kim EJ, Lee MR, Scott MB, Tweed JK, Pinloche E, Wallace RJ, Scollan ND (2011) As yet uncultured bacteria phylogenetically classified as Prevotella, Lachnospiraceae incertae sedis and unclassified Bacteroidales, Clostridiales and Ruminococcaceae may play a predominant role in ruminal biohydrogenation. Environ Microbiol 13(6):1500–1512
Julliand V, de Vaux A, Millet L, Fonty G (1999) Identification of Ruminococcus flavefaciens as the predominant cellulolytic bacterial species of the equine cecum. Appl Environ Microbiol 65(8):3738–3741
Kim M, Kim J, Kuehn L, Bono J, Berry E, Kalchayanand N, Freetly HC, Benson AK, Wells J (2014) Investigation of bacterial diversity in the feces of cattle fed different diets. J Anim Sci 92(2):683–694
Lettat A, Nozière P, Silberberg M, Morgavi D, Berger C, Martin C (2010) Experimental feed induction of ruminal lactic, propionic, or butyric acidosis in sheep. J Anim Sci 88(9):3041–3046
Lin H, An Y, Hao F, Wang Y, Tang H (2016) Correlations of fecal metabonomic and microbiomic changes induced by high-fat diet in the pre-obesity state. Sci Rep 6:21618
Mao S, Zhang R, Wang D, Zhu W (2012) The diversity of the fecal bacterial community and its relationship with the concentration of volatile fatty acids in the feces during subacute rumen acidosis in dairy cows. BMC Vet Res 8(1):237
Mao S, Zhang M, Liu J, Zhu W (2015) Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: membership and potential function. Sci Rep 5:16116
Mao SY, Huo WJ, Zhu WY (2016) Microbiome–metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ Microbiol 18(2):525–541
Miller TL, Wolin MJ (1995) Bioconversion of cellulose to acetate with pure cultures of Ruminococcus albus and a hydrogen-using acetogen. Appl Environ Microbiol 61(11):3832–3835
Owen J, Ingleton JW (1963) A study of food intake and production in grazing ewes II. The interrelationships between food intake and productive output. J Agr Sci 61(3):329–340
Qin W (1982) Determination of rumen volatile fatty acids by means of gas chromatography. J Nanjing Agric Coll 4:110–116
Sanguinetti E, Collado MC, Marrachelli VG, Monleon D, Selma-Royo M, Pardo-Tendero MM, Burchielli S, Iozzo P (2018) Microbiome-metabolome signatures in mice genetically prone to develop dementia, fed a normal or fatty diet. Sci Rep 8(1):4907
Suchodolski JS (2013) Gastrointestinal microbiota. In: Washabau RJ, Day MJ (eds) Canine & feline gastroenterology. Saunders Elsevier, St. Louis, pp 32–41
Varel V, Yen JT (1997) Microbial perspective on fiber utilization by swine. J Anim Sci 75(10):2715–2722
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73(16):5261–5267
Wang Y, Xu L, Liu J, Zhu W, Mao S (2017) A high grain diet dynamically shifted the composition of mucosa-associated microbiota and induced mucosal injuries in the colon of sheep. Front Microbiol 8:2080
Xue Y, Guo C, Hu F, Liu J, Mao S (2018) Hepatic metabolic profile reveals the adaptive mechanisms of ewes to severe undernutrition during late gestation. Metabolites 8(4):85
Xue Y, Guo C, Hu F, Sun D, Liu J, Mao S (2019a) Molecular mechanisms of lipid metabolism disorder in livers of ewes with pregnancy toxemia. Animal 13(5):992–999
Xue Y, Guo C, Hu F, Zhu W, Mao S (2019b) Maternal undernutrition induces fetal hepatic lipid metabolism disorder and affects the development of fetal liver in a sheep model. FASEB J 33(9):9990–10004
Xue Y, Guo C, Hu F, Zhu W, Mao S (2020) PPARA/RXRA signalling regulates the fate of hepatic non-esterified fatty acids in a sheep model of maternal undernutrition. BBA-Mol Cell Biol L 1865(2):158548
Ye H, Liu J, Feng P, Zhu W, Mao S (2016) Grain-rich diets altered the colonic fermentation and mucosa-associated bacterial communities and induced mucosal injuries in goats. Sci Rep 6:20329
Zheng D, Wang H-Z, Gou M, Nobu MK, Narihiro T, Hu B, Nie Y, Tang Y-Q (2019) Identification of novel potential acetate-oxidizing bacteria in thermophilic methanogenic chemostats by DNA stable isotope probing. Appl Microbiol Biotechnol 103(20):8631–8645
Funding
The present study was funded by the National Key Research and Development Program of China (grant number 2016YFD0501200).
Author information
Authors and Affiliations
Contributions
The authors’ contributions are as follows: S. M. and Y. X. conceived and designed the study; Y. X., F. H., C. G., S. M., and H. Z. conducted the research; Y. X., F. X., and S. M. analyzed and interpreted the data; Y. X. wrote the manuscript; and S. M. revised the manuscript. The authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All experimental procedures in this study were approved by the Animal Care and Use Committee of Nanjing Agricultural University (SYXK (Su) 2015–0656).
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 179 kb)
Rights and permissions
About this article
Cite this article
Xue, Y., Hu, F., Guo, C. et al. Undernutrition shifted colonic fermentation and digest-associated bacterial communities in pregnant ewes. Appl Microbiol Biotechnol 104, 5973–5984 (2020). https://doi.org/10.1007/s00253-020-10662-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10662-4