Abstract
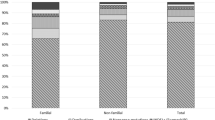
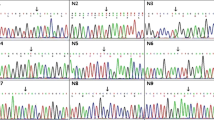
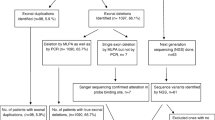
Mutations in the dystrophin gene could cause Duchenne muscular dystrophy (DMD), which is the most common muscular disorder in pediatrics. Considering the growing evidence on appropriateness of gene therapies for DMD, precise genetic diagnosis seems essential. Hence, we conducted a study to determine mutational patterns in Iranian children with DMD. To detect all probable large mutations in the dystrophin gene, 314 DMD patients were evaluated using the multiplex ligation-dependent probe amplification (MLPA). Subjects who were MLPA-negative underwent the next generation sequencing (NGS) to identify potential point mutations. MLPA detected deletions (79.93%) and duplications (5.41%) along the dystrophin gene of 268 patients. Distribution of large mutations was heterogeneous and followed hotspot pattern throughout the gene. From 46 patients who were MLPA-negative, 43 exhibited point mutations including nonsense in 7.64%, frameshifts in 4.77%, splicing in 0.96%, and missense variations in 0.32% of participants. Most of the point mutations were located between exons 19 and 40. In three patients (1%), no mutation was found using either MLPA or NGS. Two subjects had novel nonsense mutations (L1675X and E1199X) in their dystrophin gene, which were considered as the possible reason for elimination of major domains of the gene. The results of this study provided invaluable information regarding the distribution of various large and small mutations in Iranian individuals with DMD. Besides, the novel nonsense mutations L1675X and E1199X were identified within the highly conserved residues, leading to elimination of significant domains of the dystrophin gene.




Similar content being viewed by others
References
Aartsma-Rus A, Ginjaar IB, Bushby K (2016) The importance of genetic diagnosis for Duchenne muscular dystrophy. J Med Genet 53(3):145–151
Alame M, Lacourt D, Zenagui R, Mechin D, Danton F, Koenig M, Claustres M, Cossée M (2016) Implementation of a reliable next-generation sequencing strategy for molecular diagnosis of dystrophinopathies. J Mol Diagn 18(5):731–740
King N, Horrocks C, Brooks J (2018) Interviews in qualitative research. SAGE Publications Limited, London
Barzegar M, Habibi P, Bonyady M, Topchizadeh V, Shiva S (2015) Exon deletion pattern in duchene muscular dystrophy in north west of Iran. Iran J Child Neurol 9(1):42–48
Beggs AH, Koenig M, Boyce FM, Kunkel LM (1990) Detection of 98% of DMD/BMD gene deletions by polymerase chain reaction. Hum Genet 86(1):45–48
Behjati S, Tarpey PS (2013) What is next generation sequencing? Arch Dis Child Educa Pract Ed 98(6):236–238
Bladen CL, Rafferty K, Straub V, Monges S, Moresco A, Dawkins H, Roy A, Chamova T, Guergueltcheva V, Korngut L, Campbell C, Dai Y, Barišić N, Kos T, Brabec P, Rahbek J, Lahdetie J, Tuffery-Giraud S, Claustres M, Leturcq F, Ben Yaou R, Walter MC, Schreiber O, Karcagi V, Herczegfalvi A, Viswanathan V, Bayat F, de la caridad Guerrero Sarmiento I, Ambrosini A, Ceradini F, Kimura E, van den Bergen JC, Rodrigues M, Roxburgh R, Lusakowska A, Oliveira J, Santos R, Neagu E, Butoianu N, Artemieva S, Rasic VM, Posada M, Palau F, Lindvall B, Bloetzer C, Karaduman A, Topaloğlu H, Inal S, Oflazer P, Stringer A, Shatillo AV, Martin AS, Peay H, Flanigan KM, Salgado D, von Rekowski B, Lynn S, Heslop E, Gainotti S, Taruscio D, Kirschner J, Verschuuren J, Bushby K, Béroud C, Lochmüller H (2013) The TREAT-NMD Duchenne muscular dystrophy registries: conception, design, and utilization by industry and academia. Hum Mutat 34(11):1449–1457
Bladen CL, Salgado D, Monges S, Foncuberta ME, Kekou K, Kosma K, Dawkins H, Lamont L, Roy AJ, Chamova T, Guergueltcheva V, Chan S, Korngut L, Campbell C, Dai Y, Wang J, Barišić N, Brabec P, Lahdetie J, Walter MC, Schreiber-Katz O, Karcagi V, Garami M, Viswanathan V, Bayat F, Buccella F, Kimura E, Koeks Z, van den Bergen JC, Rodrigues M, Roxburgh R, Lusakowska A, Kostera-Pruszczyk A, Zimowski J, Santos R, Neagu E, Artemieva S, Rasic VM, Vojinovic D, Posada M, Bloetzer C, Jeannet PY, Joncourt F, Díaz-Manera J, Gallardo E, Karaduman AA, Topaloğlu H, el Sherif R, Stringer A, Shatillo AV, Martin AS, Peay HL, Bellgard MI, Kirschner J, Flanigan KM, Straub V, Bushby K, Verschuuren J, Aartsma-Rus A, Béroud C, Lochmüller H (2015) The TREAT-NMD DMD Global Database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum Mutat 36(4):395–402
Chen C, Ma H, Zhang F, Chen L, Xing X, Wang S, Zhang X, Luo Y (2014) Screening of Duchenne muscular dystrophy (DMD) mutations and investigating its mutational mechanism in Chinese patients. PLoS One 9(9):e108038
Cho A, Seong MW, Lim BC, Lee HJ, Byeon JH, Kim SS, Kim SY, Choi SA, Wong AL, Lee J, Kim JS, Ryu HW, Lee JS, Kim H, Hwang H, Choi JE, Kim KJ, Hwang YS, Hong KH, Park S, Cho SI, Lee SJ, Park H, Seo SH, Park SS, Chae JH (2017) Consecutive analysis of mutation spectrum in the dystrophin gene of 507 Korean boys with Duchenne/Becker muscular dystrophy in a single center. Muscle Nerve 55(5):727–734
Dardiotis E, Aloizou A-M, Siokas V, Patrinos GP, Deretzi G, Mitsias P, Aschner M, Tsatsakis A (2018) The role of microRNAs in patients with amyotrophic lateral sclerosis. J Mol Neurosci 66(4):617–628
Elhawary NA, Shawky RM, Hashem N (2004) Frameshift deletion mechanisms in Egyptian Duchenne and Becker muscular dystrophy families. Mol Cells 18(2):141–149
Flanagan SE, Patch A-M, Ellard S (2010) Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genet Test Mol Biomark 14(4):533–537
Flanigan KM, Dunn DM, von Niederhausern A, Soltanzadeh P, Gappmaier E, Howard MT, Sampson JB, Mendell JR, Wall C, King WM, Pestronk A, Florence JM, Connolly AM, Mathews KD, Stephan CM, Laubenthal KS, Wong BL, Morehart PJ, Meyer A, Finkel RS, Bonnemann CG, Medne L, Day JW, Dalton JC, Margolis MK, Hinton VJ, the United Dystrophinopathy Project Consortium, Weiss RB (2009) Mutational spectrum of DMD mutations in dystrophinopathy patients: application of modern diagnostic techniques to a large cohort. Hum Mutat 30(12):1657–1666
Foster H, Popplewell L, Dickson G (2012) Genetic therapeutic approaches for Duchenne muscular dystrophy. Hum Gene Ther 23(7):676–687
Gao QQ, McNally EM (2011) The dystrophin complex: structure, function, and implications for therapy. Compr Physiol 5(3):1223–1239
Hallwirth Pillay KD, Bill PL, Madurai S, Mubaiwa L, Rapiti P (2007) Molecular deletion patterns in Duchenne and Becker muscular dystrophy patients from KwaZulu Natal. J Neurol Sci 252(1):1–3
Hamzi K, Itto AB, Itri M, Nadifi S (2014) Prenatal diagnosis of BMD in Morocco: evolution and limits. J Mol Neurosci 52(4):459–460
Itto AB, Hamzi K, Bellayou H, Itri M, Slassi I, Nadifi S (2013) Evolution of molecular diagnosis of Duchenne muscular dystrophy. J Mol Neurosci 50(2):314–316
Iyombe-Engembe J-P, Tremblay JP (2017) The advances and challenges of gene therapy for Duchenne muscular dystrophy. J Genetic Med Gene Therapy 1:019–036
Janssen B, Hartmann C, Scholz V, Jauch A, Zschocke J (2005) MLPA analysis for the detection of deletions, duplications and complex rearrangements in the dystrophin gene: potential and pitfalls. Neurogenetics. 6(1):29–35
Juan-Mateu J, Gonzalez-Quereda L, Rodriguez MJ, Baena M, Verdura E, Nascimento A, Ortez C, Baiget M, Gallano P (2015) DMD mutations in 576 dystrophinopathy families: a step forward in genotype-phenotype correlations. PLoS One 10(8):e0135189
Khordadpoor-Deilamani F, Akbari MT, Nafissi S, Zamani G (2011) Dystrophin gene mutation analysis in Iranian males and females using multiplex polymerase chain reaction and multiplex ligation-dependent probe amplification methods. Genet Test Mol Biomark 15(12):893–899
Li Y, Liu Z, OuYang S, Zhu Y, Wang L, Wu J (2016) Distribution of dystrophin gene deletions in a Chinese population. J Int Med Res 44(1):99–108
Liang WC, Wang CH, Chou PC, Chen WZ, Jong YJ (2018) The natural history of the patients with Duchenne muscular dystrophy in Taiwan: a medical center experience. Pediatr Neonatol 59(2):176–183
Lorant J, Saury C, Schleder C, Robriquet F, Lieubeau B, Négroni E, Leroux I, Chabrand L, Viau S, Babarit C, Ledevin M, Dubreil L, Hamel A, Magot A, Thorin C, Guevel L, Delorme B, Péréon Y, Butler-Browne G, Mouly V, Rouger K (2018) Skeletal muscle regenerative potential of human MuStem cells following transplantation into injured mice muscle. Mol Ther 26(2):618–633
Ma P, Zhang S, Zhang H, Fang S, Dong Y, Zhang Y, Hao W, Wu S, Zhao Y (2018) Comprehensive genetic characteristics of dystrophinopathies in China. Orphanet J Rare Dis 13(1):109
Madania A, Zarzour H, Jarjour RA, Ghoury I (2010) Combination of conventional multiplex PCR and quantitative real-time PCR detects large rearrangements in the dystrophin gene in 59% of Syrian DMD/BMD patients. Clin Biochem 43(10–11):836–842
Manjunath M, Kiran P, Preethish-Kumar V, Nalini A, Singh RJ, Gayathri N (2015) A comparative study of mPCR, MLPA, and muscle biopsy results in a cohort of children with Duchenne muscular dystrophy: a first study. Neurol India 63(1):58–62
Mohammed F, Elshafey A, Al-Balool H, Alaboud H, Al Ben Ali M, Baqer A et al (2018) Mutation spectrum analysis of Duchenne/Becker muscular dystrophy in 68 families in Kuwait: the era of personalized medicine. PLoS One 13(5):e0197205
Muntoni F, Torelli S, Ferlini A (2003) Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol 2(12):731–740
Nouri N, Fazel-Najafabadi E, Salehi M, Hosseinzadeh M, Behnam M, Ghazavi MR et al (2014) Evaluation of multiplex ligation-dependent probe amplification analysis versus multiplex polymerase chain reaction assays in the detection of dystrophin gene rearrangements in an Iranian population subset. Adv Biomed Res 3:72
Nowak KJ, Davies KE (2004) Duchenne muscular dystrophy and dystrophin: pathogenesis and opportunities for treatment. EMBO Rep 5(9):872–876
Okubo M, Minami N, Goto K, Goto Y, Noguchi S, Mitsuhashi S, Nishino I (2016) Genetic diagnosis of Duchenne/Becker muscular dystrophy using next-generation sequencing: validation analysis of DMD mutations. J Hum Genet 61(6):483–489
Okubo M, Goto K, Komaki H, Nakamura H, Mori-Yoshimura M, Hayashi YK, Mitsuhashi S, Noguchi S, Kimura E, Nishino I (2017) Comprehensive analysis for genetic diagnosis of dystrophinopathies in Japan. Orphanet J Rare Dis 12(1):149
Perkins KJ, Davies KE (2018) Alternative utrophin mRNAs contribute to phenotypic differences between dystrophin-deficient mice and Duchenne muscular dystrophy. FEBS Lett 592(11):1856–1869
Rininsland F, Reiss J (1994) Microlesions and polymorphisms in the Duchenne/Becker muscular dystrophy gene. Hum Genet 94(2):111–116
Ryder S, Leadley RM, Armstrong N, Westwood M, de Kock S, Butt T, Jain M, Kleijnen J (2017) The burden, epidemiology, costs and treatment for Duchenne muscular dystrophy: an evidence review. Orphanet J Rare Dis 12(1):79
Sbiti A, El Kerch F, Sefiani A (2002) Analysis of dystrophin gene deletions by multiplex PCR in Moroccan patients. J Biomed Biotechnol 2(3):158–160
Schwartz M, Duno M (2004) Improved molecular diagnosis of dystrophin gene mutations using the multiplex ligation-dependent probe amplification method. Genet Test 8(4):361–367
Siokas V, Aslanidou P, Aloizou A-M, Peristeri E, Stamati P, Liampas I, et al (2020) Does the CD33 rs3865444 polymorphism confer susceptibility to Alzheimer’s disease? J Mol Neurosci 1–10
Sokratous M, Dardiotis E, Bellou E, Tsouris Z, Michalopoulou A, Dardioti M, Siokas V, Rikos D, Tsatsakis A, Kovatsi L, Bogdanos DP, Hadjigeorgiou GM (2018) CpG island methylation patterns in relapsing-remitting multiple sclerosis. J Mol Neurosci 64(3):478–484
Suh MR, Lee KA, Kim EY, Jung J, Choi WA, Kang SW (2017) Multiplex ligation-dependent probe amplification in X-linked recessive muscular dystrophy in Korean subjects. Yonsei Med J 58(3):613–618
Tennyson CN, Klamut HJ, Worton RG (1995) The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat Genet 9(2):184–190
Torella A, Trimarco A, Blanco Fdel V, Cuomo A, Aurino S, Piluso G et al (2010) One hundred twenty-one dystrophin point mutations detected from stored DNA samples by combinatorial denaturing high-performance liquid chromatography. J Mol Diagn 12(1):65–73
Tuffery-Giraud S, Beroud C, Leturcq F, Yaou RB, Hamroun D, Michel-Calemard L et al (2009) Genotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD-DMD database: a model of nationwide knowledgebase. Hum Mutat 30(6):934–945
Ulgenalp A, Giray O, Bora E, Hizli T, Kurul S, Sagin-Saylam G et al (2004) Deletion analysis and clinical correlations in patients with Xp21 linked muscular dystrophy. Turk J Pediatr 46(4):333–338
Vainzof M, Ayub-Guerrieri D, Onofre PC, Martins PC, Lopes VF, Zilberztajn D et al (2008) Animal models for genetic neuromuscular diseases. J Mol Neurosci 34(3):241–248
Wang X, Wang Z, Yan M, Huang S, Chen TJ, Zhong N (2008) Similarity of DMD gene deletion and duplication in the Chinese patients compared to global populations. Behav Brain Funct 4:20
Yamagishi H, Kato S, Hiraishi Y, Ishihara T, Hata J, Matsuo N, Takano T (1996) Identification of carriers of Duchenne/Becker muscular dystrophy by a novel method based on detection of junction fragments in the dystrophin gene. J Med Genet 33(12):1027–1031
Zamani G, Heidari M, Azizi Malamiri R, Ashrafi MR, Mohammadi M, Shervin Badv R, Hosseini SA, Salehi S, Shahrokhi A, Qorbani M, Fathi MR (2016) The quality of life in boys with Duchenne muscular dystrophy. Neuromuscul Disord 26(7):423–427
Zhang A, Uaesoontrachoon K, Shaughnessy C, Das JR, Rayavarapu S, Brown KJ, Ray PE, Nagaraju K, van den Anker JN, Hoffman EP, Hathout Y (2015) The use of urinary and kidney SILAM proteomics to monitor kidney response to high dose morpholino oligonucleotides in the mdx mouse. Toxicol Rep 2:838–849
Acknowledgments
The authors would like to thank the patients and their parents for their cooperation. We also would like to thank Ramak Heidari, Neda Anisi, and Samira Anisi, the Iranian Muscular Dystrophy Association members for their assistance in the process of data collection.
Funding
This research is supported by a grant from Tehran University of Medical Sciences (TUMS). The funding organization had no role in the design and conduct of the study; in the collection, analysis, and interpretation of the data; or in the preparation, review, or approval of the article and the decision to submit the article for publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zamani, G., Hosseini Bereshneh, A., Azizi Malamiri, R. et al. The First Comprehensive Cohort of the Duchenne Muscular Dystrophy in Iranian Population: Mutation Spectrum of 314 Patients and Identifying Two Novel Nonsense Mutations. J Mol Neurosci 70, 1565–1573 (2020). https://doi.org/10.1007/s12031-020-01594-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-020-01594-9