Abstract
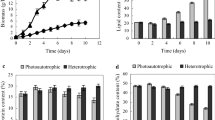
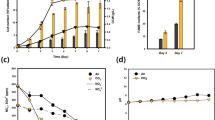
The mixed culture of microalgae and yeast has been confirmed to enhance many bioprocesses including waste degradation, remediation, and biomass generation for lipid production. Our latest study showed that besides the higher yield of biomass and lipid, the mixed culture of Chlorella pyrenoidosa and Yarrowia lipolytica can achieve the higher yields of carbohydrates, protein, and higher heating value. The strengthening mechanism of the mixed culture of microalgae and yeast mostly focused on nutritional complementarity and the culture conditions in previous works. In this work, de novo transcriptome profiling and central metabolic pathway analysis of C. pyrenoidosa cells in mono microalgae culture and mixed culture were performed. The differential transcriptome analysis revealed that most biological processes of C. pyrenoidosa, such as TCA cycle, glycolysis/gluconeogenesis pathway, pentose phosphate pathway, and nitrate assimilation, were enhanced by the mixed culture with yeast at 24 h. Meanwhile, the CO2 assimilation of C. pyrenoidosa was inhibited. Deep analysis of carbon metabolism, nitrogen metabolism, and photosynthesis well illustrates the intrinsic mechanism of physiological and biochemical differences occurring in the mono microalgae culture and the mixed culture. This study unraveled metabolic regulation molecular mechanism of C. pyrenoidosa in response to the mixed culture with Y. lipolytica, and strengthens the understanding of the synergistic effect between microalgae and yeast in the mixed culture.




Similar content being viewed by others
References
Addy MM, Kabir F, Zhang R, Lu Q, Deng X, Current D, Griffith R, Ma Y, Zhou W, Chen P, Ruan R (2017) Co-cultivation of microalgae in aquaponic systems. Bioresour Technol 245:27–34
Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60
Cheirsilp B, Suwannarat W, Niyomdecha R (2011) Mixed culture of oleaginous yeast Rhodotorula glutinis and microalga Chlorella vulgaris for lipid production from industrial wastes and its use as biodiesel feedstock. Nat Biotechnol 28:362–368
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Li L, Zhang GQ, Wang QH (2016) De novo transcriptomic analysis of Chlorella sorokiniana reveals differential genes expression in photosynthetic carbon fixation and lipid production. BMC Microbiol 16:223
Ling J, Nip S, Cheok WL, de Toledo RA, Shim H (2014) Lipid production by a mixed culture of oleaginous yeast and microalga from distillery and domestic mixed wastewater. Bioresour Technol 173:132–139
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–11
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419
Poong SW, Lim PE, Phang SM, Wong CY, Pai TW, Chen CM, Yang CH, Liu CC (2018) Transcriptome sequencing of an Antarctic microalga, Chlorella sp (Trebouxiophyceae, Chlorophyta) subjected to short-term ultraviolet radiation stress. J Appl Phycol 30:87–99
Qin L, Liu L, Zeng AP, Wei D (2017) From low-cost substrates to single cell oils synthesized by oleaginous yeasts. Bioresour Technol 245:1507–1519
Qin L, Liu L, Wang Z, Chen W, Wei D (2018a) Efficient resource recycling from liquid digestate by microalgae-yeast mixed culture and the assessment of key gene transcription related to nitrogen assimilation in microalgae. Bioresour Technol 264:90–97
Qin L, Wei D, Wang Z, Alam MA (2018b) Advantage assessment of mixed culture of Chlorella vulgaris and Yarrowia lipolytica for treatment of liquid digestate of yeast industry and cogeneration of biofuel feedstock. Appl Biochem Biotechnol 187:856–869
Qin L, Liu L, Wang Z, Chen W, Wei D (2019) The mixed culture of microalgae Chlorella pyrenoidosa and yeast Yarrowia lipolytica for microbial biomass production. Bioprocess Biosyst Eng 42:1409–1419
Rai MP, Nigam S, Sharma R (2013) Response of growth and fatty acid compositions of Chlorella pyrenoidosa under mixotrophic cultivation with acetate and glycerol for bioenergy application. Biomass Bioenergy 58:251–257
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139
Santos CA, Ferreira ME, da Silva TL, Gouveia L, Novais JM, Reis A (2011) A symbiotic gas exchange between bioreactors enhances microalgal biomass and lipid productivities: taking advantage of complementary nutritional modes. J Ind Microbiol Biotechnol 38:909–917
Sirikhachornkit A, Suttangkakul A, Vuttipongchaikij S, Juntawong P (2018) De novo transcriptome analysis and gene expression profiling of an oleaginous microalga Scenedesmus acutus TISTR8540 during nitrogen deprivation-induced lipid accumulation. Sci Rep 8:3668
Takabayashi M, Wilkerson FP, Robertson D (2005) Response of glutamine synthetase gene transcription and enzyme activity to external nitrogen sources in the diatom Skeletonema costatum (Bacillariophyceae). J Phycol 41:84–94
Tomaszewska L, Rywinska A, Gladkowski W (2012) Production of erythritol and mannitol by Yarrowia lipolytica yeast in media containing glycerol. J Ind Microbiol Biotechnol 39:1333–1343
Yamada T, Letunic I, Okuda S, Kanehisa M, Bork P (2011) iPath2.0: interactive pathway explorer. Nucleic Acids Res 39:W412–W415
Yang C, Hua Q, Shimizu K (2000) Energetics and carbon metabolism during growth of microalgal cells under photoautotrophic, mixotrophic and cyclic light-autotrophic/dark-heterotrophic conditions. Biochem Eng J 6:87–102
Yen HW, Chen PW, Chen LJ (2015) The synergistic effects for the co-cultivation of oleaginous yeast-Rhodotorula glutinis and microalgae-Scenedesmus obliquus on the biomass and total lipids accumulation. Bioresour Technol 184:148–152
Yu MJ, Yang SJ, Lin XZ (2016) De-novo assembly and characterization of Chlorella minutissima UTEX2341 transcriptome by paired-end sequencing and the identification of genes related to the biosynthesis of lipids for biodiesel. Mar Genomics 25:69–74
Zhang Z, Ji H, Gong G, Zhang X, Tan T (2014) Synergistic effects of oleaginous yeast Rhodotorula glutinis and microalga Chlorella vulgaris for enhancement of biomass and lipid yields. Bioresour Technol 164:93–99
Funding
This work was supported by the National Natural Science Foundation of China (21606230) and the Key Project of Guangdong Basic and Applied Basic Research Fund (Dongguan Joint Fund) (2019B1515120002) and the SinoPec Technology Development Program (218017-1, 36100002-19-FW2099-0035).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 1354 kb)
Rights and permissions
About this article
Cite this article
Qin, L., Wei, D. Transcriptome analysis reveals metabolic regulation mechanism of microalga Chlorella pyrenoidosa in response to the mixed culture with yeast Yarrowia lipolytica. J Appl Phycol 32, 2841–2849 (2020). https://doi.org/10.1007/s10811-020-02135-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-020-02135-y