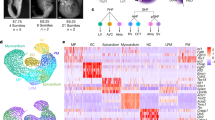
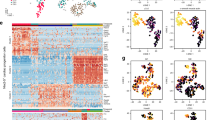
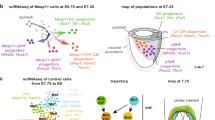
Abstract
Purpose of Review
Emerging single-cell RNA sequencing technologies hold great promises to boost our understanding of the heterogeneity and molecular regulation of diverse cell phenotypes during organ development. In this review, we aimed at summarizing recent advances in employing single-cell transcriptomic analysis to depict the landscape of embryonic heart development, in particular, focusing on cardiac progenitor (CP) differentiation.
Recent Findings
Recent studies unbiasedly cataloged and characterized cardiac cell types in the spatial and temporal resolution during early heart development. Pseudo-time analysis revealed a temporal continuum of the differentiation progress from embryonic day (E) 6.5 to E9.5, implicating early cardiac lineage restriction during mouse gastrulation. First and second heart field (FHF and SHF) CPs adopted different differentiation strategies and underwent distinct transcriptional regulation. Collectively, the comprehensive molecular atlases yield a rich resource for identification of the key cardiac regulators and signaling molecules within the key cardiac gene regulatory network (GRN) governing cardiac cell fate determinations.
Summary
This review offers insights into the exquisite process and its regulation of CP differentiation at single-cell resolution. As single-cell technologies continuously grow and evolve, computational integration of multimodal single-cell data with well-designed experimental validation promises to further delineate molecular basis in deploying cardiac progenitors of distinct sources with anatomical information.

Similar content being viewed by others
Abbreviations
- CP:
-
Cardiac progenitor
- FHF:
-
First heart field
- SHF:
-
Second heart field
- OFT:
-
Outflow tract
- LA:
-
Left atrium
- RA:
-
Right atrium
- LV:
-
Left ventricle
- RV:
-
Right ventricle
- GRN:
-
Gene regulatory network
- CM:
-
Cardiomyocyte
- EC:
-
Endothelial cell
- SMC:
-
Smooth muscle cell
- TF:
-
Transcription factor
- CHD:
-
Congenital heart disease
- KO:
-
Knock out
- WT:
-
Wild type
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6(11):826–35.
Vincent SD, Buckingham ME. How to make a heart: the origin and regulation of cardiac progenitor cells. Curr Top Dev Biol. 2010;90:1–41.
Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11(5):723–32.
Wu SM, Fujiwara Y, Cibulsky SM, Clapham DE, Lien CL, Schultheiss TM, et al. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127(6):1137–50.
Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127(6):1151–65.
Hill JT, Demarest B, Gorsi B, Smith M, Yost HJ. Heart morphogenesis gene regulatory networks revealed by temporal expression analysis. Development. 2017;144(19):3487–98.
Waardenberg AJ, Ramialison M, Bouveret R, Harvey RP. Genetic networks governing heart development. Cold Spring Harb Perspect Med. 2014;4(11):a013839.
Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126(15):3437–47.
Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566(7745):496–502.
Pijuan-Sala B, Griffiths JA, Guibentif C, Hiscock TW, Jawaid W, Calero-Nieto FJ, et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature. 2019;566(7745):490–5.
Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352(6293):1586–90.
Ellsworth DL, Blackburn HL, Shriver CD, Rabizadeh S, Soon-Shiong P, Ellsworth RE. Single-cell sequencing and tumorigenesis: improved understanding of tumor evolution and metastasis. Clin Transl Med. 2017;6(1):15.
Zhou F, Li X, Wang W, Zhu P, Zhou J, He W, et al. Tracing haematopoietic stem cell formation at single-cell resolution. Nature. 2016;533(7604):487–92.
Dulken BW, Leeman DS, Boutet SC, Hebestreit K, Brunet A. Single-cell tsranscriptomic analysis defines heterogeneity and transcriptional dynamics in the adult neural stem cell lineage. Cell Rep. 2017;18(3):777–90.
Fletcher RB, Das D, Gadye L, Street KN, Baudhuin A, Wagner A, et al. Deconstructing olfactory stem cell trajectories at single-cell resolution. Cell Stem Cell. 2017;20(6):817–30 e8.
Camp JG, Sekine K, Gerber T, Loeffler-Wirth H, Binder H, Gac M, et al. Multilineage communication regulates human liver bud development from pluripotency. Nature. 2017;546(7659):533–8.
Li L, Dong J, Yan L, Yong J, Liu X, Hu Y, et al. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell. 2017;20(6):858–73.e4.
Bondue A, Blanpain C. Mesp1: a key regulator of cardiovascular lineage commitment. Circ Res. 2010;107(12):1414–27.
Lescroart F, Chabab S, Lin X, Rulands S, Paulissen C, Rodolosse A, et al. Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat Cell Biol. 2014;16(9):829–40 This study harnesses scRNA-seq analysis to reconstruct the differentiation trajectories of earliest cardiovascular lineages.
Bruneau BG, Logan M, Davis N, Levi T, Tabin CJ, Seidman JG, et al. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt- Oram syndrome. Dev Biol. 1999;211(1):100–8.
Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5(6):877–89.
Liang X, Wang G, Lin L, Lowe J, Zhang Q, Bu L, et al. HCN4 dynamically marks the first heart field and conduction system precursors. Circ Res. 2013; 113(4):399–407.
Lescroart F, Wang X, Lin X, Swedlund B, Gargouri S, Sanchez-Danes A, et al. Defining the earliest step of cardiovascular lineage segregation by single-cell RNA-seq. Science. 2018;359(6380):1177–81.
Harvey RP. Patterning the vertebrate heart. Nat Rev Genet. 2002;3(7):544–56.
Ivanovitch K, Temino S, Torres M. Live imaging of heart tube development in mouse reveals alternating phases of cardiac differentiation and morphogenesis. Elife. 2017;6:e30668.
Xiong H, Luo Y, Yue Y, Zhang J, Ai S, Li X, et al. Single-cell transcriptomics reveals chemotaxis-mediated intraorgan crosstalk during cardiogenesis. Circ Res. 2019;125(4):398–410 This study reports that SHF CPs are guided to migrate into heart tube via MIF-CXCR2/CXCR4 signaling emanated from FHF cells.
de Soysa TY, Ranade SS, Okawa S, Ravichandran S, Huang Y, Salunga HT, et al. Single-cell analysis of cardiogenesis reveals basis for organ-level developmental defects. Nature. 2019;572(7767):120–4 This study presents a foundation of how a TF selectively regulates targeted lineages, the barrier to which leads to halted developmental progression.
DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, et al. Single-cell resolution of temporal gene expression during heart development. Dev Cell. 2016;39(4):480–90.
Xiao Y, Hill MC, Zhang M, Martin TJ, Morikawa Y, Wang SY, et al. Hippo signaling plays an essential role in cell state transitions during cardiac fibroblast development. Dev Cell. 2018;45(2):153–69.
Skelly DA, Squiers GT, McLellan MA, Bolisetty MT, Robson P, Rosenthal NA, et al. Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep. 2018;22(3):600–10.
Goodyer WR, Beyersdorf BM, Paik DT, Tian L, Li G, Buikema JW, et al. Transcriptomic profiling of the developing cardiac conduction system at single-cell resolution. Circ Res. 2019;125(4):379–97.
Li G, Xu A, Sim S, Priest JR, Tian X, Khan T, et al. Transcriptomic profiling maps anatomically patterned subpopulations among single embryonic cardiac cells. Dev Cell. 2016;39(4):491–507.
Friedman CE, Nguyen Q, Lukowski SW, Helfer A, Chiu HS, Miklas J, et al. Single-cell transcriptomic analysis of cardiac differentiation from human PSCs reveals HOPX-dependent cardiomyocyte maturation. Cell Stem Cell. 2018;23(4):586–98 e8.
Jia G, Preussner J, Chen X, Guenther S, Yuan X, Yekelchyk M, et al. Single cell RNA-seq and ATAC-seq analysis of cardiac progenitor cell transition states and lineage settlement. Nat Commun. 2018;9(1):4877 This study combines scRNA-seq and scATAC-seq analysis to understand cardiac lineage specification from both transcriptomic and epigenomic perspective.
Haghverdi L, Lun ATL, Morgan MD, Marioni JC. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol. 2018;36(5):421–7.
Cui Y, Zheng Y, Liu X, Yan L, Fan X, Yong J, et al. Single-cell transcriptome analysis maps the developmental track of the human heart. Cell Rep. 2019;26(7):1934–50 e5.
Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–502.
Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363(6434):1463–7.
Vickovic S, Eraslan G, Salmen F, Klughammer J, Stenbeck L, Schapiro D, et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat Methods. 2019;16(10):987–90.
Stahl PL, Salmen F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353(6294):78–82.
Eng CL, Lawson M, Zhu Q, Dries R, Koulena N, Takei Y, et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature. 2019;568(7751):235–9.
Asp M, Giacoomello S, Larsson L, et al. A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell. 2019;179(7):1647–60.
Bruneau BG. Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harb Perspect Biol. 2013;5(3):a008292.
Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313(5795):1922–7.
Olson EN, Srivastava D. Molecular pathways controlling heart development. Science. 1996;272(5262):671–6.
Klaus A, Muller M, Schulz H, Saga Y, Martin JF, Birchmeier W. Wnt/beta-catenin and Bmp signals control distinct sets of transcription factors in cardiac progenitor cells. Proc Natl Acad Sci U S A. 2012;109(27):10921–6.
Wang W, Niu X, Stuart T, Jullian E, Mauck WM 3rd, Kelly RG, et al. A single-cell transcriptional roadmap for cardiopharyngeal fate diversification. Nat Cell Biol. 2019;21(6):674–86 This study provides comprehensive information on cardiopharyngeal lineage segregation and origins of FHF and SHF CPs inCiona.
Francou A, Saint-Michel E, Mesbah K, Théveniau-Ruissy M, Rana MS, Christoffels VM, et al. Second heart field cardiac progenitor cells in the early mouse embryo. Biochim Biophys Acta. 2013;1833(4):795–8.
McCulley DJ, Black BL. Transcription factor pathways and congenital heart disease. Curr Top Dev Biol. 2012;100:253–77.
Behiry EG, Al-Azzouny MA, Sabry D, Behairy OG, Salem NE. Association of NKX2-5, GATA4, and TBX5 polymorphisms with congenital heart disease in Egyptian children. Mol Genet Genomic Med. 2019;7(5):e612.
Emmert-Streib F, Dehmer M, Haibe-Kains B. Gene regulatory networks and their applications: understanding biological and medical problems in terms of networks. Front Cell Dev Biol. 2014;2:38.
Thompson D, Regev A, Roy S. Comparative analysis of gene regulatory networks: from network reconstruction to evolution. Annu Rev Cell Dev Biol. 2015;31:399–428.
Scialdone A, Tanaka Y, Jawaid W, Moignard V, Wilson NK, Macaulay IC, et al. Resolving early mesoderm diversification through single-cell expression profiling. Nature. 2016;535(7611):289–93.
Kitajima S, Takagi A, Inoue T, Saga Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development. 2000;127(15):3215–26.
Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet. 1997;16(2):154–60.
Saelens W, Cannoodt R, Todorov H, Saeys Y. A comparison of single-cell trajectory inference methods. Nat Biotechnol. 2019;37(5):547–54.
Cannoodt R, Saelens W, Saeys Y. Computational methods for trajectory inference from single-cell transcriptomics. Eur J Immunol. 2016;46(11):2496–506.
Alemany A, Florescu M, Baron CS, Peterson-Maduro J, van Oudenaarden A. Whole-organism clone tracing using single-cell sequencing. Nature. 2018;556(7699):108–12.
Spanjaard B, Hu B, Mitic N, Olivares-Chauvet P, Janjuha S, Ninov N, et al. Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars. Nat Biotechnol. 2018;36(5):469–73.
Schmidt ST, Zimmerman SM, Wang J, Kim SK, Quake SR. Quantitative analysis of synthetic cell lineage tracing using nuclease barcoding. ACS Synth Biol. 2017;6(6):936–42.
McKenna A, Findlay GM, Gagnon JA, Horwitz MS, Schier AF, Shendure J. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science. 2016;353(6298):aaf7907.
Raj B, Wagner DE, McKenna A, Pandey S, Klein AM, Shendure J, et al. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat Biotechnol. 2018;36(5):442–50.
Shema E, Bernstein BE, Buenrostro JD. Single-cell and single-molecule epigenomics to uncover genome regulation at unprecedented resolution. Nat Genet. 2019;51(1):19–25.
Ai S, Xiong H, Li CC, Luo Y, Shi Q, Liu Y, et al. Profiling chromatin states using single-cell itChIP-seq. Nat Cell Biol. 2019;21(9):1164–72 This study provides the first example to demonstrate the differential transition pattern between transcriptome and enhancers of CP differentiation in single-cell resolution using both scRNA-seq and sc-itChIP-seq.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Regenerative Medicine
Rights and permissions
About this article
Cite this article
Xiong, H., He, A. Single-Cell Transcriptomic Analysis of Cardiac Progenitor Differentiation. Curr Cardiol Rep 22, 38 (2020). https://doi.org/10.1007/s11886-020-01285-2
Published:
DOI: https://doi.org/10.1007/s11886-020-01285-2