Abstract
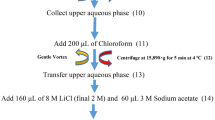
The RNA extraction was performed from foliar (F) and whole wheat plants (including rhizosphere) (WP) samples by (1) the standard TRIzol® protocol, and (2) a modified CTAB and TRIzol® protocol. The modified CTAB and TRIzol® protocol was able to extract high-quality RNA (205.96 ± 18.7 ng/µL for F, and 231.76 ± 66.8 ng/µL for WP; RIN > 8.0), compared to the standard TRIzol® protocol (92.73 ± 24.2 ng/µL for F, and WP completely degraded, RIN < 8.0). Real-time RT-PCR assay was carried out for 6-SFT1 (target) and 18S rRNA (housekeeping) genes, which showed a PCR efficiency of 111% and 118%, respectively, and a no significant relative expression (3773 ± 1383.8 for F and 2847 ± 1037.5 for WP) was observed from RNA extracted by the modified protocol. The modified CTAB and TRIzol® protocol was able to produce high-quality RNA (yield, purity, and integrity) from foliar and whole wheat plants (including rhizosphere with recalcitrant properties).




Similar content being viewed by others
References
Aali KA, Parsinejad M, Rahmani B (2009) Estimation of saturation percentage of soil using multiple regression, ANN, and ANFIS techniques. Comput Inf Sci 2(3):127–136
Alm EW, Zheng D, Raskin L (2000) The presence of humic substances and DNA in RNA extracts affects hybridization results. Appl Environ Microbiol 66(10):4547–4554
Byrnes S, Fan A, Trueb J, Jareczek F, Mazzochette M, Sharon A, Sauer-Budge AF, Klapperich CM (2013) A portable, pressure driven, room temperature nucleic acid extraction and storage system for point of care molecular diagnostics. Anal Methods 5(13):3177–3184
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal Biochem 162(1):156–159
Chomczynski P, Sacchi N (2006) The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: twenty-something years on. Nat Protoc 1(2):581–585
Fischer M, Renevey N, Thür B, Hoffmann D, Beer M, Hoffmann B (2016) Efficacy assessment of nucleic acid decontamination reagents used in molecular diagnostic laboratories. PLoS ONE 11(7):1–9
Franchi M, Ferris JP, Gallori E (2003) Cations as mediators of the adsorption of nucleic acids on clay surfaces in prebiotic environments. Orig Life Evol Biosph 33(1):1–16
Gambino G, Perrone I, Gribaudo I (2008) A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem Anal 19(6):520–525
Jordon-Thaden IE, Chanderbali AS, Gitzendanner MA, Soltis DE (2015) Modified CTAB and TRIzol protocols improve RNA extraction from chemically complex Embryophyta. Appl in Plant Sci. https://doi.org/10.3732/apps.1400105
Knief C (2014) Analysis of plant microbe interactions in the era of next generation sequencing technologies. Front Plant Sci 5:1–23
Lehmann J, Kleber M (2015) The contentious nature of soil organic matter. Nature 528(7580):60–68
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408
Matlock B (2015) Assessment of nucleic acid purity—NanoDrop spectrophotometers. In: ThermoFisher Scientific. https://www.thermoscientific.com. Accessed 19 Dec 2019
Mueller O, Lightfoot S, Schröder A (2004) RNA Integrity Number (RIN) Standardization of RNA Quality Control. Tech Rep 5989-1165EN, Agilent Technologies, Application Note. http://www.agilent.com/chem/labonachip
Nielsen H (ed) (2011) Working with RNA. In: RNA. Methods in Molecular Biology (Methods and Protocols), vol 703. Humana Press, Totowa. https://doi.org/10.1007/978-1-59745-248-9_2
Novinscak A, Filion M (2011) Effect of soil clay content on RNA isolation and on detection and quantification of bacterial gene transcripts in soil by quantitative reverse Transcription-PCR. Appl Environ Microbiol 77(17):6249–6252
Pedreira-Segade U, Hao J, Razafitianamaharavo A, Pelletier M, Marry V, Le Crom S, Michot L, Daniel I (2018) How do nucleotides adsorb onto clays? Life 8(4):59
Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Menzel W, Granzow M, Ragg T (2006) The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 7:1–14
Valenzuela-Aragon B, Parra-Cota FI, Santoyo G, Arellano-Wattenbarger GL, de los Santos-Villalobos S (2019) Plant-assisted selection: a promising alternative for in vivo identification of wheat (Triticum turgidum L. subsp. Durum) growth promoting bacteria. Plant Soil 435(1):367–384. https://doi.org/10.1007/s11104-018-03901-1
Verhulst N, Deckers J, Govaerts B (2009) Classification of the soil at CIMMYT ’ s experimental station in the Yaqui Valley near Ciudad Obregón, Sonora, México. In CIMMYT Report. https://repository.cimmyt.org/xmlui/bitstream/handle/10883/562/94513.pdf. Accessed 1 Jan 2020
Villa-Rodríguez E, Ibarra-Gámez C, de los Santos-Villalobos S (2018) Extraction of high-quality RNA from Bacillus subtilis with a lysozyme pre-treatment followed by the Trizol method. J Microbiol Methods. https://doi.org/10.1016/j.mimet.2018.02.011
Wang Y, Fujii T (2011) Evaluation of methods of determining humic acids in nucleic acid samples for molecular biological analysis. Biosci Biotechnol Biochem 75(2):355–357
Wang Y, Hayatsu M, Fujii T (2012) Extraction of bacterial RNA from soil: challenges and solutions. Microbes Environ 27(2):111–121
Wilfinger WW, Mackey K, Chomczynski P (1997) Effect of pH and ionic strength on the spectrophotometric assessment of nucleic acid purity. Biotechniques 22(3):474–481
Wong LM, Silvaraj S, Phoon LQ (2015) An optimised high-salt CTAB protocol for both DNA and RNA isolation from succulent stems of Hylocereus sp. J Med Bioeng 3(4):236–240
Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14(6):415–421
Zipper H (2003) Mechanisms underlying the impact of humic acids on DNA quantification by SYBR Green I and consequences for the analysis of soils and aquatic sediments. Nucl Acids Res 31(7):39e–39
Acknowledgements
The authors acknowledge support by the CONACyT Project 257246 “Interacción trigo x microorganismos promotores del crecimiento vegetal: identificando genes con potencial agro-biotecnológico”, and the Instituto Tecnológico de Sonora (ITSON) Project PROFAPI 2020_0013 “Bacillus sp. TSO9: afiliación taxonómica a nivel del genoma e identificación de genes asociados a la promoción del crecimiento en el trigo”. Luis Abraham Chaparro-Encinas and Guillermo Luis Arellano-Wattenbarger were supported by CONACYT fellowships 292582 and 626633, respectively.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Posta.
Rights and permissions
About this article
Cite this article
Chaparro-Encinas, L.A., Arellano-Wattenbarger, G.L., Parra-Cota, F.I. et al. A modified CTAB and Trizol® protocol for high-quality RNA extraction from whole wheat seedlings, including rhizosphere. CEREAL RESEARCH COMMUNICATIONS 48, 275–282 (2020). https://doi.org/10.1007/s42976-020-00046-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42976-020-00046-9