Abstract
To accomplish the remarkable task of lifelong infection, the Epstein–Barr virus (EBV) switches between four viral genome latency and lytic programmes to navigate the B-cell compartment and evade immune responses. The transforming programme, consisting of highly immunogenic EBV nuclear antigen (EBNA) and latent membrane proteins (LMPs), is expressed in newly infected B lymphocytes and in post-transplant lymphomas. On memory cell differentiation and in most EBV-associated Burkitt’s lymphomas, all but one viral antigen are repressed for immunoevasion. To gain insights into the epigenetic mechanisms that restrict immunogenic oncoprotein expression, a genome-scale CRISPR–Cas9 screen was performed in EBV and Burkitt’s lymphoma cells. Here, we show that the ubiquitin ligase ubiquitin-like PHD and RING finger domain-containing protein 1 (UHRF1) and its DNA methyltransferase partner DNA methyltransferase I (DNMT1) are critical for the restriction of EBNA and LMP expression. All UHRF1 reader and writer domains were necessary for silencing and DNMT3B was identified as an upstream viral genome CpG methylation initiator. Polycomb repressive complex I exerted a further layer of control over LMP expression, suggesting a second mechanism for latency programme switching. UHRF1, DNMT1 and DNMT3B are upregulated in germinal centre B cells, the Burkitt’s lymphoma cell of origin, providing a molecular link between B-cell state and the EBV latency programme. These results suggest rational therapeutic targets to manipulate EBV oncoprotein expression.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The CRISPR screen results are available in the Source Data for Fig. 1. The accession numbers for the RNA-seq datasets reported in this paper are Gene Expression Omnibus (GEO) nos. GSE136596, GSE136597 and GSE136609. Published microarray analysis53 of human tonsil B-cell naïve, germinal centre, memory or plasma cell subsets were also used and are available under GEO accession number GSE24919. The data that support the findings of this study are available from the corresponding author upon request. Source Data for Figs. 1–6 and Extended Data Figs. 1, 2 and 4–10 are provided with the paper.
Change history
03 May 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41564-022-01137-5
References
Longnecker, R. M., Kieff, E. & Cohen, J. I. in Fields Virology 6th edn, Vol. 1 (Wolters Kluwer Health Adis (ESP), 2013).
Cohen, J. I. Epstein–Barr virus infection. N. Engl. J. Med. 343, 481–492 (2000).
Thorley-Lawson, D. A. EBV persistence–introducing the virus. Curr. Top. Microbiol. Immunol. 390, 151–209 (2015).
Price, A. M. & Luftig, M. A. To be or not IIb: a multi-step process for Epstein–Barr virus latency establishment and consequences for B cell tumorigenesis. PLoS Pathog. 11, e1004656 (2015).
Woellmer, A. & Hammerschmidt, W. Epstein–Barr virus and host cell methylation: regulation of latency, replication and virus reactivation. Curr. Opin. Virol. 3, 260–265 (2013).
Price, A. M., Messinger, J. & Luftig, M. A. c-Myc represses transcription of Epstein–Barr virus latent membrane protein 1 early after primary B cell infection. J. Virol. 92, e01178-17 (2018).
Mrozek-Gorska, P. et al. Epstein–Barr virus reprograms human B lymphocytes immediately in the prelatent phase of infection. Proc. Natl Acad. Sci. USA 116, 16046–16055 (2019).
Wang, L. W. et al. Epstein–Barr-virus-induced one-carbon metabolism drives B cell transformation. Cell Metab. 30, 539–555 (2019).
Seifert, M., Scholtysik, R. & Kuppers, R. Origin and pathogenesis of B cell lymphomas. Methods Mol. Biol. 971, 1–25 (2013).
Westhoff Smith, D. & Sugden, B. Potential cellular functions of Epstein–Barr nuclear antigen 1 (EBNA1) of Epstein–Barr virus. Viruses 5, 226–240 (2013).
Sugden, B. Epstein–Barr virus: the path from association to causality for a ubiquitous human pathogen. PLoS Biol. 12, e1001939 (2014).
Schaefer, B. C., Strominger, J. L. & Speck, S. H. Host-cell-determined methylation of specific Epstein–Barr virus promoters regulates the choice between distinct viral latency programs. Mol. Cell. Biol. 17, 364–377 (1997).
Masucci, M. G. et al. 5-Azacytidine up regulates the expression of Epstein–Barr virus nuclear antigen 2 (EBNA-2) through EBNA-6 and latent membrane protein in the Burkitt’s lymphoma line rael. J. Virol. 63, 3135–3141 (1989).
Kalla, M., Schmeinck, A., Bergbauer, M., Pich, D. & Hammerschmidt, W. AP-1 homolog BZLF1 of Epstein–Barr virus has two essential functions dependent on the epigenetic state of the viral genome. Proc. Natl Acad. Sci. USA 107, 850–855 (2010).
Robertson, K. D., Hayward, S. D., Ling, P. D., Samid, D. & Ambinder, R. F. Transcriptional activation of the Epstein–Barr virus latency C promoter after 5-azacytidine treatment: evidence that demethylation at a single CpG site is crucial. Mol. Cell. Biol. 15, 6150–6159 (1995).
Niller, H. H., Szenthe, K. & Minarovits, J. Epstein–Barr virus–host cell interactions: an epigenetic dialog? Front. Genet. 5, 367 (2014).
Hughes, D. J. et al. Contributions of CTCF and DNA methyltransferases DNMT1 and DNMT3B to Epstein–Barr virus restricted latency. J. Virol. 86, 1034–1045 (2012).
Gregory, C. D., Rowe, M. & Rickinson, A. B. Different Epstein–Barr virus-B cell interactions in phenotypically distinct clones of a Burkitt’s lymphoma cell line. J. Gen. Virol. 71, 1481–1495 (1990).
Babcock, G. J., Hochberg, D. & Thorley-Lawson, D. A. The expression pattern of Epstein–Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity 13, 497–506 (2000).
Carter, K. L., Cahir-McFarland, E. & Kieff, E. Epstein–Barr virus-induced changes in B-lymphocyte gene expression. J. Virol. 76, 10427–10436 (2002).
Peng, M. & Lundgren, E. Transient expression of the Epstein–Barr virus LMP1 gene in B-cell chronic lymphocytic leukemia cells, T cells, and hematopoietic cell lines: cell-type-independent-induction of CD23, CD21, and ICAM-1. Leukemia 7, 104–112 (1993).
Kis, L. L., Takahara, M., Nagy, N., Klein, G. & Klein, E. IL-10 can induce the expression of EBV-encoded latent membrane protein-1 (LMP-1) in the absence of EBNA-2 in B lymphocytes and in Burkitt lymphoma- and NK lymphoma-derived cell lines. Blood 107, 2928–2935 (2006).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR–Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Bostick, M. et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764 (2007).
Sharif, J. et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912 (2007).
Avvakumov, G. V. et al. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455, 822–825 (2008).
Bashtrykov, P., Jankevicius, G., Jurkowska, R. Z., Ragozin, S. & Jeltsch, A. The UHRF1 protein stimulates the activity and specificity of the maintenance DNA methyltransferase DNMT1 by an allosteric mechanism. J. Biol. Chem. 289, 4106–4115 (2014).
Chen, C. et al. Uhrf1 regulates germinal center B cell expansion and affinity maturation to control viral infection. J. Exp. Med. 215, 1437–1448 (2018).
Shaknovich, R. et al. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation. Blood 118, 3559–3569 (2011).
Wang, L. W., Jiang, S. & Gewurz, B. E. Epstein–Barr virus LMP1-mediated oncogenicity. J. Virol. 91, e01718-16 (2017).
Kieser, A. & Sterz, K. R. The latent membrane protein 1 (LMP1). Curr. Top. Microbiol. Immunol. 391, 119–149 (2015).
Wang, C. et al. RNA sequencing analyses of gene expression during Epstein–Barr virus infection of primary B lymphocytes. J. Virol. 93, e00226-19 (2019).
Price, A. M. et al. Analysis of Epstein–Barr virus-regulated host gene expression changes through primary B-cell outgrowth reveals delayed kinetics of latent membrane protein 1-mediated NF-κB activation. J. Virol. 86, 11096–11106 (2012).
Minamitani, T. et al. Mouse model of Epstein–Barr virus LMP1- and LMP2A-driven germinal center B-cell lymphoproliferative disease. Proc. Natl Acad. Sci. USA 114, 4751–4756 (2017).
Cohen, J. I., Wang, F., Mannick, J. & Kieff, E. Epstein–Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc. Natl Acad. Sci. USA 86, 9558–9562 (1989).
Pei, Y., Banerjee, S., Jha, H. C., Sun, Z. & Robertson, E. S. An essential EBV latent antigen 3C binds Bcl6 for targeted degradation and cell proliferation. PLoS Pathog. 13, e1006500 (2017).
Vaughan, R. M. et al. Chromatin structure and its chemical modifications regulate the ubiquitin ligase substrate selectivity of UHRF1. Proc. Natl Acad. Sci. USA 115, 8775–8780 (2018).
Maenohara, S. et al. Role of UHRF1 in de novo DNA methylation in oocytes and maintenance methylation in preimplantation embryos. PLoS Genet. 13, e1007042 (2017).
Bronner, C., Alhosin, M., Hamiche, A. & Mousli, M. Coordinated dialogue between UHRF1 and DNMT1 to ensure faithful inheritance of methylated DNA patterns. Genes (Basel) 10, 65 (2019).
Cheng, J. et al. Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein. J. Biol. Chem. 288, 1329–1339 (2013).
Arita, K. et al. Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc. Natl Acad. Sci. USA 109, 12950–12955 (2012).
Harrison, J. S. et al. Hemi-methylated DNA regulates DNA methylation inheritance through allosteric activation of H3 ubiquitylation by UHRF1. eLife 5, e17101 (2016).
Robertson, K. D. et al. CpG methylation of the major Epstein–Barr virus latency promoter in Burkitt’s lymphoma and Hodgkin’s disease. Blood 88, 3129–3136 (1996).
Kretzmer, H. et al. DNA methylome analysis in Burkitt and follicular lymphomas identifies differentially methylated regions linked to somatic mutation and transcriptional control. Nat. Genet. 47, 1316–1325 (2015).
Bhende, P. M., Seaman, W. T., Delecluse, H.-J. & Kenney, S. C. The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat. Genet. 36, 1099–1104 (2004).
Bergbauer, M. et al. CpG-methylation regulates a class of Epstein–Barr virus promoters. PLoS Pathog. 6, e1001114 (2010).
Smits, A. H. et al. Biological plasticity rescues target activity in CRISPR knock outs. Nat. Methods 16, 1087–1093 (2019).
Giffin, L. & Damania, B. KSHV: pathways to tumorigenesis and persistent infection. Adv. Virus Res. 88, 111–159 (2014).
Lurain, K. et al. Viral, immunologic, and clinical features of primary effusion lymphoma. Blood 133, 1753–1761 (2019).
Anastasiadou, E. et al. Epigenetic mechanisms do not control viral latency III in primary effusion lymphoma cells infected with a recombinant Epstein–Barr virus. Leukemia 19, 1854–1856 (2005).
Goll, M. G. & Bestor, T. H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514 (2005).
Barwick, B. G. et al. B cell activation and plasma cell differentiation are inhibited by de novo DNA methylation. Nat. Commun. 9, 1900 (2018).
Lai, A. Y. et al. DNA methylation profiling in human B cells reveals immune regulatory elements and epigenetic plasticity at Alu elements during B-cell activation. Genome Res. 23, 2030–2041 (2013).
Leonard, S. et al. Epigenetic and transcriptional changes which follow Epstein–Barr virus infection of germinal center B cells and their relevance to the pathogenesis of Hodgkin’s lymphoma. J. Virol. 85, 9568–9577 (2011).
Kelly, G., Bell, A. & Rickinson, A. Epstein–Barr virus-associated Burkitt lymphomagenesis selects for downregulation of the nuclear antigen EBNA2. Nat. Med. 8, 1098–1104 (2002).
Allday, M. J. EBV finds a polycomb-mediated, epigenetic solution to the problem of oncogenic stress responses triggered by infection. Front. Genet. 4, 212 (2013).
Arvey, A. et al. An atlas of the Epstein–Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe 12, 233–245 (2012).
Sterlin, D. et al. Genetic, cellular and clinical features of ICF syndrome: a French national survey. J. Clin. Immunol. 36, 149–159 (2016).
Salamon, D. et al. Protein-DNA binding and CpG methylation at nucleotide resolution of latency-associated promoters Qp, Cp, and LMP1p of Epstein–Barr virus. J. Virol. 75, 2584–2596 (2001).
Simon, J. A. & Kingston, R. E. Occupying chromatin: polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49, 808–824 (2013).
Taylor, G. S., Long, H. M., Brooks, J. M., Rickinson, A. B. & Hislop, A. D. The immunology of Epstein–Barr virus-induced disease. Annu. Rev. Immunol. 33, 787–821 (2015).
Ahmed, M. et al. TCR-mimic bispecific antibodies targeting LMP2A show potent activity against EBV malignancies. JCI Insight 3, e97805 (2018).
Ma, Y. et al. CRISPR/Cas9 screens reveal Epstein–Barr virus-transformed B cell host dependency factors. Cell Host Microbe 21, 580–591 (2017).
Lu, F. et al. Coordinate regulation of TET2 and EBNA2 controls the DNA methylation state of latent Epstein–Barr virus. J. Virol. 91, e00804-17 (2017).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013).
Acknowledgements
This work was supported by National Institutes of Health grant nos. RO1s AI137337 and CA228700 (B.E.G.) and CA047006 (B.Z.), the Starr Cancer Foundation Project no. I11-0043 (L.G.R., B.E.G., E.C. and R.J.O.), the Burroughs Wellcome Career Award in Medical Sciences and the American Cancer Society Research Scholar Award (B.E.G.), the Leukemia and Lymphoma Society SCOR grant (E.C. and L.G.R.) and the Next Generation Fund at the Broad Institute (J.G.D.). We thank J. Sample for the KEM I, MUTU I and MUTU III cell lines. We thank R. Reynolds, T. Dalton, L. Kottyan, C. Forney, M. Weirauch, P. Wade and A. Chicoine for technical and bioinformatics assistance.
Author information
Authors and Affiliations
Contributions
R.G. and B.E.G. designed the screen with input from J.G.D., C.J., E.C., L.G.R. and R.J.O. R.G. performed the screen. R.G. and Y.Z. performed and analysed most of the CRISPR and biochemical experiments with assistance from M.S. The bioinformatics analysis was performed by R.G., M.T., J.G.D. and B.Z. B.E.G. supervised the study.
Corresponding author
Ethics declarations
Competing interests
L.G.R. is a consultant for Janssen and ADC Therapeutics. All other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Genome-wide CRISPR-Cas9 Mediated Loss-of-Function Screen Identifies UHRF1 and DNMT1 as candidate regulators of EBV Latency III.
a, Volcano plot visualization of -Log10(p-value) statistical significance (y-axis) and Log2 fold-change in mRNA abundance (x-axis) of MUTU III versus MUTU I cells from triplicate RNA-seq datasets. ICAM-1 and CD10 (encoded by MME) are highlighted by red dots. n = 3 biologically independent replicates. P value and log fold change were generated with DESeq under default settings with Wald test and normal shrinkage, respectively. b, Plasma membrane ICAM-1 (left) and CD10 (right) abundances in MUTU1 (red), MUTU III (green) and GM12878 LCLs. Plots are representative of n = 3 biologically independent values. c, FACS plots of plasma membrane ICAM-1 and CD10 abundances in MUTU I cells prior to and at 8 days following transduction by the Brunello sgRNA library. Transduced cells were puromycin selected two days post-transduction. Plots are representative of n = 3 biologically independent values. d, Volcano plot of CRISPR screen results for genes encoding host epigenetic factors in the ICAM-1high/CD10low and ICAM-1high/CD10high subpopulations, using the curated EpiFactors database69. Plots are representative of n = 3 biologically independent values. P-values were determined by one-sided Fisher’s exact test. e, Log2-normalized abundance of the four Brunello library sgRNAs targeting UHRF1 or DNMT1 in the input (pre-FACSort) library (gray), ICAM-1high/CD10 low population (black), ICAM-low/CD10low (light blue), and ICAM-1high/CD10high (dark blue). Significance between the input and FACSorted groups was assessed using the two-sided unpaired Student’s t test. Mean + SD from n = 3 biologically independent screens is shown.
Extended Data Fig. 2 Dynamic primary B-cell UHRF1 and DNMT1 regulation by physiological or EBV stimuli.
a, Immunoblot of WCL from peripheral blood primary human B-cells that were resting or that were stimulated by Mega-CD40L (50 ng/ml), anti-IgM (1 μg/ml), CpG (0.5 μM), IgM+ CpG or CD40 + CpG for 24 hours. b, Normalized DNMT3B, DNMT3A, DNMT1 or UHRF1 mRNA levels in primary human peripheral blood B-cells at the indicated day post infection (DPI) by the EBV B95.8 strain32. Shown are the mean + SEM values from n = 3 of biologically independent RNAseq datasets. c, UHRF1, DNMT1, DNMT3A and TET2 relative protein abundances detected by tandem-mass-tag-based proteomics at rest and at nine time points after EBV B95.8 infection of primary human peripheral blood B-cells at a multiplicity of infection of 0.1. DNMT3B expression was not detected. Data represent the average ± SEM for four independent replicates8. For each protein, the maximum level detected across the time course was set to a value of one. d, Volcano plot visualization of -Log10 (p-value) statistical significance (y-axis) and Log2 fold-change in mRNA abundance (x-axis) of ex vivo GC B-cells that conditionally expressed control GFP or both LMP1 and LMP2A from an AICDA promoter, which is activated in GC B-cells. Data are from n = 3 of biologically independent RNA-seq datasets34. P-value and log fold change were generated with DESeq under default settings with Wald test and Normal shrinkage, respectively. UHRF1, DNMT1, DNMT3A and DNMT3B values are highlighted. e, Normalized EBNA1 mRNA reads from RNAseq analysis of MUTU I (n = 3) or Rael cells (n = 2) that expressed control or UHRF1 sgRNAs, as indicated. P values were calculated by unpaired two-sided student’s t-test with equal variance assumption. f, Immunoblot analysis of WCL from MUTU III or from MUTU I that expressed the indicated control or UHRF1 sgRNAs. Cells were harvested at the indicated day post sgRNA delivery by lentivirus transduction. Blots in a and f are representative of n = 3 biologically independent replicates.
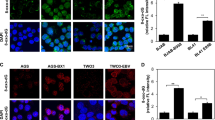
Extended Data Fig. 3 UHRF1 is necessary for silencing of latency III antigen expression.
a, Confocal immunofluoresence analysis of LMP1 or LMP2A in Rael cells with control or UHRF1 sgRNAs. Nuclei are counterstained with Hoechst 33342. Scale bar, 10 µm. Images are representative of n = 3 biologically independent experiments. b, Confocal immunofluoresence analysis of EBNA2 in Rael cells with control or UHRF1 sgRNAs. Nuclei are counterstained with Hoechst 33342. Scale bar, 10 µm. Images are representative of n = 3 biologically independent experiments.
Extended Data Fig. 4 UHRF1 depletion induces latency III and induces ICAM-1 in EBV+ but not EBV- B-cells.
a, Volcano plot visualization of -Log10 (p-value) statistical significance (y-axis) and Log2 (fold-change in mRNA abundance) in Rael cells that expressed control or UHRF1 sgRNAs. Data are from n = 2 of biologically independent RNA-seq datasets. P value and log fold change were generated with DESeq under default settings with Wald test and Normal shrinkage, respectively. Values for selected genes upregulated (red) or downregulated (blue) by UHRF1 sgRNA are highlighted. b, Enrichr pathway analysis of gene sets significantly upregulated in MUTU III versus MUTU I (gray bars), or by UHRF1 versus control sgRNA expression in MUTU I (black bars) or Rael (blue bars). Shown are the adjusted p-values from Enrich analysis of triplicate RNAseq datasets using Fisher exact test. See also Source Data for Extended Data Fig. 4. c, Heatmap representation of abundances of mRNAs encoding latency III antigens in Rael BL with control or UHRF1 sgRNAs. Shown are data from n = 2 biologically independent replicates. d, FACS analysis of total cell ICAM-1 and LMP1 expression in Rael cells with control (left) or UHRF1 sgRNA #2 (right). e, Immunoblot analysis of WCL from EBV- (left) or EBV+ Akata BLs with the indicated sgRNA expression. f, FACS analysis of plasma membrane ICAM-1 expression in the indicated cell lines with control or UHRF1 sgRNA #2 expression. KEM I and Rael are EBV+ BL. REH is an EBV-negative B-cell acute lymphoblastic leukemia cell line. Shown below are immunoblots of WCL from KEM I BLs with the indicated sgRNA expression. All FACS plots and blots are representative of n = 3 biologically independent replicates.
Extended Data Fig. 5 MeDIP analysis of promoter CpG methylation in LCLs or in CRISPR edited BL treated with acyclovir.
a. 5 mC MeDIP was performed on DNA from GM12878 (black bars), GM11830 (gray bars) or GM12881 LCLs (blue bars) followed by qPCR for the Qp, Cp, Wp or LMP1p. Mean ± SEM for n = 3 of biologically independent replicates are shown. P values were calculated using two-way ANOVA with Turkey’s multiple comparisons test. b, FACS plots of PM gp350 and ICAM-1 abundances in MUTU I cells that expressed the indicated sgRNAs and that were treated with acyclovir (50 μg/ml) to block new EBV genome synthesis by the viral polymerase. The indicated populations from sgControl versus sgUHRF1 expressing cells were sorted and used for 5 mC MeDIP-qPCR analysis. n = 3 biologically independent experiments. c, 5 mC MeDIP was performed on chromatin from sorted sgControl (black bars) or sgUHRF1 (gray bars) followed by qPCR for Cp, Wp or Qp. Mean ± SEM for n = 3 biologically independent replicates are shown. p-values were calculated by unpaired two-sided student’s t-test with equal variance assumption. d, 5 mC MeDIP was performed on chromatin from sorted sgControl (black bars) or sgUHRF1 (gray bars) followed by qPCR for the host STAT3 or TRAF1 promoters (n = 3 of biologically independent experiments). Shown also are normalized mRNA reads from n = 3 of biologically independent RNAseq datasets of MUTU I expressing control or UHRF1 sgRNAs. P values were calculated by unpaired two-sided student’s t-test with equal variance assumption.
Extended Data Fig. 6 UHRF1 depletion induced lytic reactivation in a small percentage of cells.
a, FACS analysis of ICAM-1 and gp350 plasma membrane abundances in MUTU I cells with control or UHRF1 sgRNAs. DN = double negative. Plots are representative of n = 3 biologically independent replicates. b, Immunoblot analysis of WCL from the indicated populations of FACSorted MUTU I cells, representative of n = 3 biologically independent replicates. c, Heatmap of EBV gene expression in the indicated FACSorted MUTU I populations. Heatmap values displace z-score values of mRNAs of EBV lytic genes, which describe the standard deviation variation from the mean value of each gene. Individual data from n = 3 biologically independent samples are shown. d, Principal component (PC) analysis of mRNA expression in sorted MUTU I populations. RNAseq of n = 3 biologically independent replicates was performed. e–g 5 mC MeDIP was performed on chromatin isolated from MUTU I cells followed by qPCR using primers specific for Zp (e), Qp (f), or Wp (g). Mean ± SEM are shown for n = 3 biologically independent replicates. P values were calculated using one-way ANOVA with Sidak’s multiple comparisons test.
Extended Data Fig. 7 Effects of UHRF1 or DNMT1 depletion in EBV+ primary effusion lymphoma, gastric carcinoma or nasopharyngeal carcinoma cells.
a, Immunoblot analysis of WCL from EBV+ and KSHV+ primary effusion cell line JSC-1 that expressed the indicated control, UHRF1 or DNMT1 sgRNAs or from MUTU III WCL as a control. b, 5 mC MeDIP analysis of chromatin from JSC-1 expressing control or UHRF1 sgRNA #2 followed by qPCR using primers specific for Cp or LMP1p. Mean ± SEM are shown for n = 3 biologically independent replicates. P values were calculated by unpaired two-sided student’s t-test with equal variance assumption. c, Immunoblot analysis of WCL from EBV+ NUGC3 gastric carcinoma cells that expressed the indicated sgRNAs. Quantification of EBNA2:GAPDH and LMP1:GAPDH ratios are indicated. d, Immunoblot analysis of WCL from EBV+ C666.1 nasopharyngeal carcinoma (NPC) cells that expressed the indicated control or UHRF1 sgRNAs or from MUTU III as a control. e, Immunoblot analysis of WCL from EBV+ C666.1 nasopharyngeal carcinoma (NPC) cells that expressed the indicated control or DNMT1 sgRNAs or from MUTU III as a control. f, 5mC MeDIP analysis of chromatin from C666.1 cells that expressed sgControl (gray boxes) or sgUHRF1 (black boxes) followed by qPCR using primers specific for Cp or LMP1p. Mean ± SEM are shown for n = 3 biologically independent replicates. P values were calculated by unpaired two-sided student’s t-test with equal variance assumption. Blots in a, c, d, and e are representative of n = 3 biologically independent replicates.
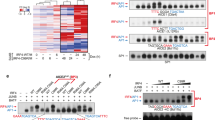
Extended Data Fig. 8 Initiator DNA methyltransferase expression in human tonsil B-cell subsets and overexpression effects on LCL growth and survival.
a. DNMT3A and 3B mRNA expression in published microarray analysis of human tonsil B-cell naïve, GC, memory or plasma cell subsets53. b, Propidium iodide (PI) cell cycle analysis of GM12878 LCLs that expressed control GFP, DNMT3A or DNMT3B. Data are representative of n = 2 biologically independent replicates. c, Percentages of the indicated cell populations in GM12878 that expressed GFP, DNMT3A or DNMT3B. Data are the average of n = 2 biologically independent replicates. d, qPCR analysis of LMP1 mRNA abundances in GM12878 that expressed the indicated GFP, DNMT3A or DNMT3B cDNAs. Mean ± SEM from n = 3 biologically independent replicates are shown. P values were calculated using one-way ANOVA with Turkey’s multiple comparisons test. e, Immunoblot analysis of WCL from GM11830 LCLs that expressed GFP, DNMT3A or DNMT3B cDNAs, as indicated. f, Immunoblot analysis of WCL from MUTU III that expressed GFP, DNMT3A or DNMT3B cDNAs. g, Normalized MUTU III live cell numbers at the indicated timepoints after expression of GFP, DNMT3A or DNMT3B. Mean ± SEM from n = 3 biologically independent replicates are shown. P values were calculated by two-sided student’s t-test with equal variance assumption. h, 5 mC MeDIP analysis of chromatin from GM12878 expressing GFP (gray box), DNMT3A (blue box) or DNMT3B (black box) followed by qPCR using a second primer set specific for LMP1p (from the same experiment as in Fig. 4f). Mean ± SEM are shown for n = 3 biologically independent replicates. p-values were calculated using two-way ANOVA with Sidak's multiple comparisons test. i, Immunoblot analysis of WCL from MUTU I, from the EBV+/KSHV+ PELs JSC-1 or BC-1, or from the EBV-/KSHV+ PEL BCBL-1. Blots in e, f, and i are representative of n = 3 biologically independent replicates.
Extended Data Fig. 9 PRC1 is a BL host factor important for silencing of LMP1 and LMP2a expression.
a, Volcano plots showing the -Log10 (p-value) statistical significance (y-axis) and Log2 fold-change (x-axis) calculated by STARS analysis of sgRNA abundance in the input versus FACSorted ICAM-1high/CD10low population. Values for genes encoding the PRC1 subunits SCML2 and RING1 are highlighted in blue, and values for genes encoding UHRF1 and DNMT1 are shown in purple for comparison. n = 3 of biologically independent replicates. P values were determined by one-sided Fisher’s exact test. b, Mean +/- SEM normalized SCML2 mRNA reads from triplicate RNAseq analysis of MUTU I versus III (left) or of MUTU I that expressed control or UHRF1 sgRNA (right). n = 3 of biologically independent replicates. P values were calculated by unpaired two-sided student’s t-test with equal variance assumption. c, Immunoblot analysis of WCL from MUTU III or from MUTU1 that expressed the indicated sgRNAs. d, qPCR analysis of EBNA3C expression. Values were normalized by 18s rRNA qPCR levels and the ratio in control cells was set to 1. Mean ± SEM from n = 3 biologically independent replicates are shown. p-values were calculated using one-way ANOVA with Sidak’s multiple comparisons. e, Immunoblot analysis of WCL from MUTU III or Daudi that expressed the indicated sgRNAs. f, Fold change of the LMP1:GAPDH immunoblot ratio (left) or BCL6:GAPDH immunoblot ratio (right) from n = 3 of biologically independent replicates, as in ED Fig. 9e. p-values were calculated using one-way ANOVA with Sidak’s multiple comparisons. g, Immunoblot analysis of WCL from MUTU III or from REH acute lymphoblastic leukemia EBV-negative B-cells or from Akata EBV-negative BL expressing the indicated sgRNAs. h, Immunoblot analysis of WCL from MUTU I that expressed the indicated sgRNAs. i, Normalized IRF4 mRNA reads from n = 3 of biologically independent triplicate RNAseq datasets of MUTU I cells that expressed sgControl or sgUHRF1. p-values were calculated by unpaired two-sided student’s t-test with equal variance assumption. Blots in c, e, g and h are representative of at least 3 biologically independent replicates.
Extended Data Fig. 10 PRC1 effects on latency III expression and EBV H2Ak119 monoubiquitylation.
a, Immunoblot analysis of WCL from GM12878 LCLs that expressed GFP versus RING1 cDNAs or from MUTU I or III as controls. b, FACS analysis of PM abundances of LMP1/NF-kB targets Fas/CD95 (left) and ICAM-1 (right) in GM12878 LCL expressing GFP (blue) versus RING1 (red). c, ChIP analysis of chromatin from MUTU 1 (black bar) or MUTU III (blue bar) using control IgG versus anti-H2A lysine 119 monoubiquitin (H2AK119Ub) followed by qPCR with primers specific for LMP1p, LMP2p or Cp. Mean ± SEM from n = 3 biologically independent replicates are shown, p-values were calculated by unpaired two-sided student’s t-test with equal variance assumption. d, Schematic model of PRC1 complex-mediated ubiquitination of chromatin. a and b are representative of n = 3 biologically independent replicates.
Supplementary information
Supplementary Information
List of primers and sgRNAs.
Source data
Source Data Fig. 1
Statistical Source Data.
Source Data Fig. 2
Statistical Source Data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical Source Data.
Source Data Fig. 4
Statistical Source Data.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Statistical Source Data.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Statistical Source Data.
Source Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical Source Data.
Source Data Extended Data Fig. 2
Statistical Source Data.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical Source Data.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 5
Statistical Source Data.
Source Data Extended Data Fig. 6
Statistical Source Data.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 7
Statistical Source Data.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 8
Statistical Source Data.
Source Data Extended Data Fig. 8
Unprocessed western blots.
Source Data Extended Data Fig. 9
Statistical Source Data.
Source Data Extended Data Fig. 9
Unprocessed western blots.
Source Data Extended Data Fig. 10
Statistical Source Data.
Source Data Extended Data Fig. 10
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Guo, R., Zhang, Y., Teng, M. et al. DNA methylation enzymes and PRC1 restrict B-cell Epstein–Barr virus oncoprotein expression. Nat Microbiol 5, 1051–1063 (2020). https://doi.org/10.1038/s41564-020-0724-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-020-0724-y
This article is cited by
-
Epigenetic alterations and advancement of lymphoma treatment
Annals of Hematology (2024)
-
Shared and distinct interactions of type 1 and type 2 Epstein-Barr Nuclear Antigen 2 with the human genome
BMC Genomics (2024)
-
Establishment and comprehensive analysis of a new human cell line (NK-NJ) with NK-cell characteristics established from extranodal natural killer cell lymphoma/leukemia
Human Cell (2022)