Abstract
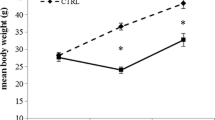
The tissue distribution pattern of lipid is highly diverse among different fish species. Tiger puffer has a special lipid storage pattern, storing lipid predominantly in liver. In order to better understand the lipid physiology in fish storing lipid in liver, the present study preliminarily investigated the tissue distribution of transcription for 29 lipid metabolism-related genes in tiger puffer, which are involved in lipogenesis, fatty acid oxidation, biosynthesis and hydrolysis of glycerides, lipid transport, and relevant transcription regulation. Samples of eight tissues, brain, eye, heart, spleen, liver, intestine, skin, and muscle, from fifteen juvenile tiger puffer were used in the qRT-PCR analysis. The intestine and brain had high transcription of lipogenic genes, whereas the liver and muscle had low expression levels. The intestine also had the highest transcription level of most apolipoproteins and lipid metabolism-related transcription factors. The transcription of fatty acid β-oxidation-related genes was low in the muscle. The peroxisomal fatty acid oxidation may dominate over mitochondrial β-oxidation in the liver and intestine of tiger puffer, and the MAG pathway probably predominates over the G3P pathway in re-acylation of absorbed lipids in the intestine. The intracellular glyceridases were highly transcribed in the brain, eye, and heart. In conclusion, in tiger puffer, the intestine could be a center of lipid metabolism whereas the liver is more likely a pure storage organ for lipid. The lipid metabolism in the muscle could also be inactive, possibly due to the very low level of intramuscular lipid.











Similar content being viewed by others
Abbreviations
- acacβ :
-
Acetyl-CoA carboxylase beta
- fas :
-
Fatty acid synthase
- scd1 :
-
Delta-9-desaturase 1
- acox1 :
-
Acyl-CoA oxidase 1, palmitoyl
- acox3 :
-
Acyl-CoA oxidase 3, pristanoyl
- cpt-1 :
-
Carnitine O-palmitoyltransferase-1
- gpat :
-
Glycerol-3-phosphate acyltransferase
- dgat1:
-
Diacylglycerol O-acyltransferase 1
- bsal :
-
Bile acid-activated lipase
- hsl :
-
Hormone-sensitive lipase
- daglα :
-
Diacylglycerol lipase, alpha
- mgll :
-
Monoglyceride lipase
- lpl :
-
Lipoprotein lipase
- lipc :
-
Lipase, hepatic
- apo :
-
Apolipoprotein
- mttp :
-
Microsomal triglyceride transfer protein
- srebf1 :
-
Sterol regulatory element binding transcription factor 1
- ppar :
-
Peroxisome proliferators-activated receptor
- fxr :
-
Farnesoid X receptor (nuclear receptor subfamily 1, group H, member 4, NR1H4)
- lxrα:
-
Liver X receptor alpha (nuclear receptor subfamily 1, group H, member 3, NR1H3)
- hnf4α :
-
Hepatocyte nuclear factor 4, alpha
- lrh-1 :
-
Liver receptor homolog-1 (nuclear receptor subfamily 5, group A, member 2, NR5A2)
- gys2 :
-
Glycogen synthase 2
References
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250. https://doi.org/10.1158/0008-5472.CAN-04-0496
Aparicio S, Chapman J, Stupka E, Putnam N, Chia J, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, Sollewijn Gelpke MD, Roach J, Oh T, Ho IY, Wong M, Detter C, Verhoef F, Predki P, Tay A, Lucas S, Richardson P, Smith SF, Clark MS, Edwards YJK, Doggett N, Zharkikh A, Tavtigian SV, Pruss D, Barnstead M, Evans C, Baden H, Powell J, Glusman G, Rowen L, Hood L, Tan YH, Elgar G, Hawkins T, Venkatesh B, Rokhsar D, Brenner S (2002) Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297(5585):1301–1310. https://doi.org/10.1126/science.1072104
Ayisi CL, Yamei C, Zhao J-L (2018) Genes, transcription factors and enzymes involved in lipid metabolism in fin fish. Agri Gene 7:7–14. https://doi.org/10.1016/j.aggene.2017.09.006
Bell MV, Batty RS, Dick JR, Fretwell K, Navarro JC, Sargent JR (1995) Dietary deficiency of docosahexaenoic acid impairs vision at lowlight intensities in juvenile herring (Clupea harengus L.). Lipids 30:443–449. https://doi.org/10.1007/BF02536303
Benítez-Santana T, Masuda R, Carrillo EJ, Ganuza E, Valencia A, Hernández-Cruz CM, Izquierdo MS (2007) Dietary n-3 HUFA deficiency induces a reduced visual response in gilthead seabream Sparus aurata larvae. Aquaculture 264:408–417. https://doi.org/10.1016/j.aquaculture.2006.10.024
Betancor MB, Sprague M, Sayanova O, Usher S, Campbell PJ, Napier JA, Caballero MJ, Tocher DR (2015) Evaluation of a high-EPA oil from transgenic Camelina sativa in feeds for Atlantic salmon (Salmo salar L.): effects on tissue fatty acid composition, histology and gene expression. Aquaculture 444:1–12. https://doi.org/10.1016/j.aquaculture.2015.03.020
Chapman MJ (1986) Comparative analysis of mammalian plasma lipoproteins. Methods Enzymol 128:70–143. https://doi.org/10.1016/0076-6879(86)28063-5
Claudel T, Staels B, Kuipers F (2005) The farnesoid X receptor: a molecular link between bile acid and lipid and glucose metabolism. Arterioscler Thromb Vasc Biol 25:2020–2030. https://doi.org/10.1161/01.ATV.0000178994.21828.a7
Collewet G, Bugeon J, Idier J, Quellec S, Quittet B, Cambert M, Haffray P (2013) Rapid quantification of muscle fat content and subcutaneous adipose tissue in fish using MRI. Food Chem 138:2008–2015. https://doi.org/10.1016/j.foodchem.2012.09.131
Crockett EL, Sidell BD (1993) Substrate selectivities differ for hepatic mitochondrial and peroxisomal β-oxidation in an Antarctic fish, Notothenia gibberifrons. Biochem J 289:427–433. https://doi.org/10.1042/bj2890427
Cruz-Garcia L, Minghetti M, Navarro I, Tocher DR (2009) Molecular cloning, tissue expression and regulation of liver X Receptor (LXR) transcription factors of Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss). Comp Biochem Physiol B 153:81–88. https://doi.org/10.1016/j.cbpb.2009.02.001
Deberardinis RJ, Lum JJ, Thompson CB (2006) Phosphatidylinositol 3-kinase-dependent modulation of carnitine palmitoyltransferase 1A expression regulates lipid metabolism during hematopoietic cell growth. J Biol Chem. 281:37372–37380. https://doi.org/10.1074/jbc.M608372200
Di Filippo M, Crehalet H, Samson-Bouma ME, Bonnet V, Aggerbeck LP, Rabes JP, Gottrand F, Luc G, Bozon D, Sassolas A (2012) Molecular and functional analysis of two new MTTP gene mutations in an atypical case of abetalipoproteinemia. J Lipid Res 53:548–555. https://doi.org/10.1194/jlr.M020024
Dong X, Xu H, Mai K, Xu W, Zhang Y, Ai Q (2015) Cloning and characterization of SREBP-1 and PPAR-α in Japanese seabass Lateolabrax japonicus, and their gene expressions in response to different dietary fatty acid profiles. Comp Biochem Physiol B 180:48–56. https://doi.org/10.1016/j.cbpb.2014.10.001
Elmerot C, Arnason U, Gojobori T, Janke A (2002) The mitochondrial genome of the pufferfish, Fugu rubripes, and ordinal teleostean relationships. Gene 295(2):163–172. https://doi.org/10.1016/s0378-1119(02)00688-1
Frøyland L, Lie Ø, Berge RK (2000) Mitochondrial and peroxisomal beta-oxidation capacities in various tissues from Atlantic salmon Salmo salar. Aquacult Nutr 6:85–89. https://doi.org/10.1046/j.1365-2095.2000.00130.x
Furuita H, Takeuchi T (1998) Effects of eicosapentaenoic and docosahexaenoic acids on growth, survival and brain development of larval Japanese flounder Paralichthys olivaceus. Aquaculture 161:269–279. https://doi.org/10.1016/S0044-8486(97)00275-5
Furukawa F, Irachi S, Koyama M, Baba O, Akimoto H, Okumura S, Kagawa H, Uchida K (2018) Changes in glycogen concentration and gene expression levels of glycogen metabolizing enzymes in muscle and liver of developing masu salmon. Comp Biochem Physiol A 225:74–82. https://doi.org/10.1016/j.cbpa.2018.07.003
Geay F, Santigosa I, Culi E, Corporeau C, Boudry PB, Dreano Y, Corcos L, Bodin N, Vandeputte M, Zambonino-Infante JL, Mazurais D, Cahu CL (2010) Regulation of FADS2 expression and activity in European sea bass (Dicentrarchus labrax, L.) fed a vegetable diet. Comp Biochem Physiol B 156:237–243. https://doi.org/10.1016/j.cbpb.2010.03.008
Gee JR, Ding Q, Keller JN (2005) Modulation of apolipoprotein E and interleukin-1beta in the aging liver. Exp Gerontol 40:409–415. https://doi.org/10.1016/j.exger.2005.01.010
Geurden I, Reyes OS, Bergot P, Coutteau P, Sorgeloos P (1998) Incorporation of fatty acids from dietary neutral lipid in eye, brain and muscle of postlarval turbot fed diets with different types of phosphatidylcholine. Fish Physiol Biochem 19:365–375. https://doi.org/10.1023/a:1007771431134
Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ (2001) Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 21:1393–1403. https://doi.org/10.1128/MCB.21.4.1393-1403.2001
He A, Ning L, Chen L, Chen Y, Xing Q, Li J, Qiao F, Li D, Zhang M, Du Z (2015) Systemic adaptation of lipid metabolism in response to low and high-fat diet in Nile tilapia (Oreochromis niloticus). Physiol Rep 3(8):e12485. https://doi.org/10.14814/phy2.12485
Horton J (2002) Sterol regulatory element-binding proteins: transcriptional activators of lipid synthesis. Biochem Soc Trans 30:1091–1095. https://doi.org/10.1042/bst0301091
Hu Y, Zheng L, Wang Q (2010) Regulation of cholesterol homeostasis by liver X receptors. Clin Chim Acta 411:617–625. https://doi.org/10.1016/j.cca.2009.12.027
Hummasti S, Tontonoz P (2006) The peroxisome proliferator-activated receptor N-terminal domain controls isotype-selective gene expression and adipogenesis. Mol Endocrinol 20(6):1261–1275. https://doi.org/10.1210/me.2006-0025
Ishizaki Y, Masuda R, Uematsu K, Shimizu K, Arimoto M, Takeuchi T (2001a) The effect of dietary docosahexaenoic acid on schooling behaviour and brain development in larval yellowtail. J Fish Biol 58:1691–1703. https://doi.org/10.1111/j.1095-8649.2001.tb02323.x
Ishizaki Y, Uematsu K, Takeuchi T (2001b) Preliminary study of the effect of dietary docosahexaenoic acid on the volumetric growth of the brain in larval yellowtail. Fisheries Sci 66:611–613. https://doi.org/10.1046/j.1444-2906.2000.00096.x
Jayakumar A, Tai M, Huang W, Al-Feel W, Hsu M, Abu-Elheiga L, Chirala SS, Wakil SJ (1995) Human fatty acid synthase: properties and molecular cloning. Proc Natl Acad Sci USA 92:8695–8699. https://doi.org/10.1073/pnas.92.19.8695
Johnston JM (1977) Gastrointestinal tissue. In: Snyder F (ed) Lipid metabolism in mammals, vol 1. Plenum Press, New York, pp 151–187
Joseph SB, Castrillo A, Laffite BA, Mangelsdorf DJ, Tontonoz P (2003) Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat Med 9:213–222. https://doi.org/10.1038/nm820
Jubie S, Ramesh PN, Dhanabal P, Kalirajan R, Muruganantham N, Antony AS (2012) Synthesis, antidepressant and antimicrobial activities of some novel stearic acid analogues. Eur J Med Chem 54:931–935. https://doi.org/10.1016/j.ejmech.2012.06.025
Kamalam BS, Panserat S, Aguirre P, Geurden I, Fontagné-Dicharry S, Médale F (2013) Selection for high muscle fat in rainbow trout induces potentially higher chylomicron synthesis and PUFA biosynthesis in the intestine. Comp Biochem Physiol A 164:417–427. https://doi.org/10.1016/j.cbpa.2012.11.020
Kaneko G, Yamada T, Han Y, Hirano Y, Khieokhajonkhet A, Shirakami H, Nagasaka R, Kondo H, Hirono I, Ushio H, Watabe S (2013) Differences in lipid distribution and expression of peroxisome proliferator-activated receptor gamma and lipoprotein lipase genes in torafugu and red seabream. Gen Comp Endocrinol 184:51–60. https://doi.org/10.1016/j.ygcen.2013.01.003
Kersten S (2014) Integrated physiology and systems biology of PPARα. Mol Metab 3:354–371. https://doi.org/10.1016/j.molmet.2014.02.002
Kikuchi K, Furuta T, Iwata N, Onuki K, Noguchi T, Sugita H (2011) Effect of dietary fatty acid composition on the growth of the tiger puffer Takifugu rubripes. Fish Sci 77:829–837. https://doi.org/10.1007/s12562-011-0393-0
Lehner R, Kuksis A (1996) Biosynthesis of triacylglycerols. Prog Lipid Res 35:169–201. https://doi.org/10.1016/0163-7827(96)00005-7
Li C, Gan F, Chen X, Liu Z, Li L, Wei W, Tang Y (2011) Molecular and expression analysis of apolipoprotein E gene in the Chinese sturgeon, Acipenser sinensis. Comp Biochem Physiol 158(B):64–70. https://doi.org/10.1016/j.cbpb.2010.09.008
Li S, Sang C, Zhang J, Chen N, Li Z, Jin P, Huang X (2018) Effects of acute hyperglycemia stress on plasma glucose, glycogen content, and expressions of glycogen synthase and phosphorylase in hybrid grouper (Epinephelus fuscoguttatus ♀ × E. lanceolatus ♂). Fish Physiol Biochem 44:1185–1196. https://doi.org/10.1007/s10695-018-0508-y
Li Y, Pang Y, Xiang X, Du J, Mai K, Ai Q (2019) Molecular cloning, characterization, and nutritional regulation of Elovl6 in large yellow croaker (Larimichthys crocea). Int J Mol Sci 20:1081. https://doi.org/10.3390/ijms20071801
Liao Z, Xu H, Wei Y, Zhang Q, Liang M (2018) Dietary astaxanthin differentially affected the lipid accumulation in the liver and muscle of the marine teleost, tiger puffer Takifugu rubripes. Aquac Res 49:3421–3433. https://doi.org/10.1111/are.13806
Liao Z, Sun B, Zhang Q, Jia L, Wei Y, Liang M, Xu H (2020) Dietary bile acids regulate the hepatic lipid homeostasis in tiger puffer fed normal or high-lipid diets. Aquaculture 519:734935. https://doi.org/10.1016/j.aquaculture.2020.734935
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001
Llewellyn L, Ramsurn VP, Wigham T, Sweeney GE, Power DM (1998) Cloning, characterisation and expression of the apolipoprotein A-I gene1 in the sea bream (Sparus aurata). Biochim Biophys Acta 1442:399–404. https://doi.org/10.1016/S0167-4781(98)00171-7
Lu K-L, Wang L-N, Zhang D-D, Liu W-B, Xu W-N (2017) Berberine attenuates oxidative stress and hepatocytes apoptosis via protecting mitochondria in blunt snout bream Megalobrama amblycephala fed high-fat diets. Fish Physiol Biochem 43:65–76. https://doi.org/10.1007/s10695-016-0268-5
Mohd-Yusof NY, Monroig O, Mohd-Adnan A, Wan KL, Tocher DR (2010) Investigation of highly unsaturated fatty acid metabolism in the Asian sea bass, Lates calcarifer. Fish Physiol Biochem 36:827–843. https://doi.org/10.1007/s10695-010-9409-4
Noffs MD, Martino RC, Trugo LC, Urbinati EC, Fernandes JBK, Takahashi LS (2009) Dietary fish oil replacement with lard and soybean oil affects triacylglycerol and phospholipid muscle and liver docosahexaenoic acid content but not in the brain and eyes of surubim juveniles Pseudoplatystoma sp. Fish Physiol Biochem 35:399–412. https://doi.org/10.1007/s10695-008-9264-8
Oaxaca-Castillo D, Andreoletti P, Vluggens A, Yu S, van Veldhoven PP, Reddy JK, Cherkaoui-Malki M (2007) Biochemical characterization of two functional human liver acyl-CoA oxidase isoforms 1a and 1b encoded by a single gene. Biochem Biophys Res Commun 360:314–319. https://doi.org/10.1016/j.bbrc.2007.06.059
Ohta K, Miyamoto H, Yaguchi T, Nagai K, Yamamoto S, Nomura T (2003) Stearic acid facilitates hippocampal neurotransmission by enhancing nicotinic ACh receptor responses via a PKC pathway. Mol Brain Res 119:83–89. https://doi.org/10.1016/j.molbrainres.2003.08.017
Olsen RE, Ringo E (1997) Lipid digestibility in fish: a review. In: Pandalai SG (ed) Recent research development in lipids Research, vol 1. Research Signpost, Irvine, pp 199–265
Oxley A, Torstensen BE, Rustan AC, Olsen RE (2005) Enzyme activities of intestinal triacylglycerol and phosphatidylcholine biosynthesis in Atlantic salmon (Salmo salar L.). Comp Biochem Physiol B 141:77–87. https://doi.org/10.1016/j.cbpc.2005.01.012
Oxley A, Jutfelt F, Sundell K, Olsen RE (2007) Sn-2-monoacylglycerol, not glycerol, is preferentially utilised for triacylglycerol and phosphatidylcholine biosynthesis in Atlantic salmon (Salmo salar L.) intestine. Comp Biochem Physiol B 146:115–123. https://doi.org/10.1016/j.cbpb.2006.09.007
Pierron F, Baudrimont M, Bossy A, Bourdineaud JP, Brethes D, Elie P, Massabuau JC (2007) Impairment of lipid storage by cadmium in the European eel (Anguilla anguilla). Aquat Toxicol 81:304–311. https://doi.org/10.1016/j.aquatox.2006.12.014
Roman-Padilla J, Rodríguez-Rúa A, Carballo C, Manchado M, Sakamoto A, Kawasaki T, Kazawa T, Ohashi R, Jiang S, Maejima T, Tanaka T, Iwanari H, Hamakubo T, Sakai J, Kodama T, Naito M (2007) Expression of liver X Receptor α in rat fetal tissues at different development stages. J Histochem Cytochem 55:641–649. https://doi.org/10.1369/jhc.6A7120.2007
Roman-Padilla J, Rodríguez-Rua A, Claros MG, Hachero-Cruzado I, Manchado M (2016) Genomic characterization and expression analysis of four apolipoprotein A-IV paralogs in Senegalese sole (Solea senegalensis Kaup). Comp Biochem Physiol B 191:84–98. https://doi.org/10.1016/j.cbpb.2015.09.010
Shi X, Sun J, Yang Z, Li X, Ji H, Li Y, Chang Z, Du Z, Chen L (2017) Molecular characterization and nutritional regulation of carnitine palmitoyltransferase (CPT) family in grass carp (Ctenopharyngodon idellus). Comp Biochem Physiol B 203:11–19. https://doi.org/10.1016/j.cbpb.2016.08.006
Tiku P, Gracey A, Macartney A, Beynon R, Cossins A (1996) Cold-induced expression of Δ9-desaturase in carp by transcriptional and posttranslational mechanisms. Science 271:815–818. https://doi.org/10.1126/science.271.5250.815
Tocher DR (2003) Metabolism and functions of lipids and fatty acids in teleost fish. Rev Fish Sci 11:107–184. https://doi.org/10.1080/713610925
Tocher DR, Fonseca-Madrigal J, Bell JG, Dick JR, Henderson RJ, Sargent JR (2002) Effects of diets containing linseed oil on fatty acid desaturation and oxidation in hepatocytes and intestinal enterocytes in Atlantic salmon (Salmo salar). Fish Physiol Biochem 26:157–170. https://doi.org/10.1023/A:1025416731014
Tocher DR, Fonseca-Madrigal J, Dick JR, Ng W-K, Bell JG, Campbell PJ (2004) Effects of water temperature and diets containing palm oil on fatty acid desaturation and oxidation in hepatocytes and intestinal enterocytes of rainbow trout (Oncorhynchus mykiss). Comp Biochem Physiol B 137:49–63. https://doi.org/10.1016/j.cbpc.2003.10.002
Tocher DR, Zheng X, Schlechtriem C, Hastings N, Dick JR, Teale AJ (2006) Highly unsaturated fatty acid synthesis in marine fish: cloning, functional characterization, and nutritional regulation of fatty acyl Δ6 desaturase of Atlantic cod (Gadus morhua L.). Lipids 41:1003–1016. https://doi.org/10.1007/s11745-006-5051-4
Toussaint C, Fauconneau B, Médale F, Collewet G, Akoka S, Haffray P, Davenel A (2005) Description of the heterogeneity of lipid distribution in the flesh of brown trout (Salmo trutta) by MR imaging. Aquaculture 243:255–267. https://doi.org/10.1016/j.aquaculture.2004.09.029
Turchetto-Zolet AC, Maraschin FS, de Morais GL, Cagliari A, Andrade CM, Margis-Pinheiro M, Margis R (2011) Evolutionary view of acyl-CoA diacylglycerol acyltransferase (DGAT), a key enzyme in neutral lipid biosynthesis. BMC Evol Biol 11:263. https://doi.org/10.1186/1471-2148-11-263
Vandesompele J, De PK, Pattyn F, Poppe B, Van RN, De PA, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034. https://doi.org/10.1186/gb-2002-3-7-research0034
Walczak R, Tontonoz P (2002) PPARadigms and PPARadoxes: expanding roles for PPARγ in the control of lipid metabolism. J Lipid Res 43:177–186. https://doi.org/10.1089/107999002753536248
Wang H, Du J, Lu S, Yao Y, Hunter F, Black DD (2001) Regulation of intestinal apolipoprotein A-I synthesis by dietary phosphatidylcholine in newborn swine. Lipids 36(7):683–687. https://doi.org/10.1007/s11745-001-0773-x
Wang Z, Li G, Nie B, Lu Y, Yin M (2006) Neuroprotective effect of the stearic acid against oxidative stress via phosphatidylinositol 3-kinase pathway. Chem-Biol Interact 160:80–87. https://doi.org/10.1016/j.cbi.2005.12.008
Wang B, Liu W, Xu C, Cao X, Zhong X, Shi H, Li X (2017) Dietary carbohydrate levels and lipid sources modulate the growth performance, fatty acid profiles and intermediary metabolism of blunt snout bream Megalobrama amblycephala in an interactive pattern. Aquaculture 481:140–153. https://doi.org/10.1016/j.aquaculture.2017.08.034
Wei J, Gao P, Zhang P, Guo Ma XM, Wei S, Yan Y, Qin Q (2015) Isolation and function analysis of apolipoprotein A-I gene response to virus infection in grouper. Fish Shellfish Immunol 43:396–404. https://doi.org/10.1016/j.fsi.2015.01.006
Wu J, Zhang J, Du X, Shen Y, Lao X, Zhang M, Chen L, Du Z (2015) Evaluation of the distribution of adipose tissues in fish using magnetic resonance imaging (MRI). Aquaculture 448:112–122. https://doi.org/10.1016/j.aquaculture.2015.06.002
Wu K, Zheng J, Luo Z, Chen Q, Zhu Q, Hu W (2016) Carnitine palmitoyltransferase I gene in Synechogobius hasta: cloning, mRNA expression and transcriptional regulation by insulin in vitro. Gene 576:429–440. https://doi.org/10.1016/j.gene.2015.10.055
Xu W, Liu W, Lu K, Jiang Y, Li G (2012) Effect of trichlorfon on oxidative stress and hepatocyte apoptosis of Carassius auratus gibelio in vivo. Fish Physiol Biochem 38:769–775. https://doi.org/10.1007/s10695-011-9559-z
Xu H, Dong X, Ai Q, Mai K, Xu W, Zhang Y, Zuo R (2014) Regulation of tissue LC-PUFA contents, Δ6 fatty acyl desaturase (FADS2) gene expression and the methylation of the putative FADS2 gene promoter by different dietary fatty acid profiles in Japanese seabass (Lateolabrax japonicus). PLoS One 9:e87726. https://doi.org/10.1371/journal.pone.0087726
Xu H, Zhang D, Yu D, Lv C, Luo H, Wang Z (2015) Molecular cloning and expression analysis of scd1 gene from large yellow croaker Larimichthys crocea under cold stress. Gene 568:100–108. https://doi.org/10.1016/j.gene.2015.05.027
Xu H, Liao Z, Zhang Q, Wei Y, Liang M (2019a) A moderately high level of dietary lipid inhibited the protein secretion function of liver in juvenile tiger puffer Takifugu rubripes. Aquaculture 498:17–27. https://doi.org/10.1016/j.aquaculture.2018.08.033
Xu H, Liao Z, Zhang Q, Wei Y, Liang M (2019b) Effects of dietary n-6 polyunsaturated fatty acids on growth performance, body composition, haematological parameters and hepatic physiology of juvenile tiger puffer (Takifugu rubripes). Aquacult Nutr 25:1073–1086. https://doi.org/10.1111/anu.12924
Xu H, Zhang Q, Wei Y, Liao Z, Liang M (2019c) Dietary methionine increased the lipid accumulation in juvenile tiger puffer Takifugu rubripes. Comp Biochem Physiol B 230:19–28. https://doi.org/10.1016/j.cbpb.2019.01.005
Xu H, Zhang Q, Shin-Kwon K, Liao Z, Wei Y, Sun B, Jia L, Chi S, Liang M (2020) Dietary taurine stimulates the hepatic biosynthesis of both bile acid and cholesterol in the marine teleost, tiger puffer (Takifugu rubripes). Br J Nutr In press. https://doi.org/10.1017/S0007114520000161
You C, Jiang D, Zhang Q, Xie D, Wang S, Dong Y, Li Y (2017) Cloning and expression characterization of peroxisome proliferator-activated receptors (PPARs) with their agonists, dietary lipids, and ambient salinity in rabbitfish Siganus canaliculatus. Comp Biochem Physiol B 206:54–64. https://doi.org/10.1016/j.cbpb.2017.01.005
Yu G, Ou W, Liao Z, Xu H, Liang M, Zhang Y, Mai K (2019) Intestinal homeostasis of juvenile tiger puffer Takifugu rubripes was sensitive to dietary arachidonic acid in terms of mucosal barrier and microbiota. Aquaculture 502:97–106. https://doi.org/10.1016/j.aquaculture.2018.12.020
Yuan D, Wu Z, Wang Y (2016) Evolution of the diacylglycerol lipases. Prog Lipid Res 64:85–97. https://doi.org/10.1016/j.plipres.2016.08.004
Zhang T, Yu F, Guo L, Chen M, Yuan X, Wu B (2018) Small heterodimer partner regulates circadian cytochromes p450 and drug-induced hepatotoxicity. Theranostics 8:5246–5258. https://doi.org/10.7150/thno.28676
Zheng JL (2013) Molecular characterization, tissue distribution and kinetic analysis of carnitine palmitoyltransferease I in juvenile yellow catfish Pelteobagrus fulvidraco. Genomics 101:195–203. https://doi.org/10.1016/j.ygeno.2012.12.002
Zheng X, Ding Z, Xu Y, Monroig O, Morais S (2009) Physiological roles of fatty acyl desaturases and elongases in marine fish: Characterisation of cDNAs of fatty acyl Δ6 desaturase and elovl5 elongase of cobia (Rachycentron canadum). Aquaculture 290:122–131. https://doi.org/10.1016/j.aquaculture.2009.02.010
Zhou WH, Rahimnejad S, Lu K, Wang L, Liu W (2019) Effects of berberine on growth, liver histology, and expression of lipid-related genes in blunt snout bream (Megalobrama amblycephala) fed high-fat diets. Fish Physiol Biochem 45:83–91. https://doi.org/10.1007/s10695-018-0536-7
Funding
This work was supported by the Central Public-interest Scientific Institution Basal Research Fund, CAFS (2018HY-ZD0505, 2020TD48), the National Key R&D Program of China (2018YFD0900400), the National Natural Science Foundation of China (31972803), and China Agriculture Research System (CARS-47-G15).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xu, H., Meng, X., Jia, L. et al. Tissue distribution of transcription for 29 lipid metabolism-related genes in Takifugu rubripes, a marine teleost storing lipid predominantly in liver. Fish Physiol Biochem 46, 1603–1619 (2020). https://doi.org/10.1007/s10695-020-00815-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10695-020-00815-7