Abstract
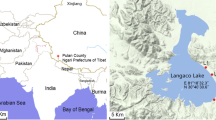
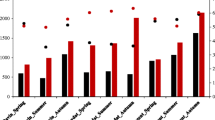
Puma Yumco Lake (PYL) is an ultraoligotrophic freshwater lake that sits an altitude of 5030 m within the Qinghai-Tibet Plateau of China. The bacterial and archaeal diversity of the lake remains poorly understood, despite their potential to inform on biogeochemical cycling and environment-microbial associations in these unique environments. Here, the bacterial and archaeal communities of PYL were investigated using high-throughput sequencing analysis of community 16S rRNA gene sequences. Further, the relationships among dominant taxa and environmental factors were comprehensively evaluated. Bacterial diversity comprised 31 phyla and 371 genera (10,645 operational taxonomic units [OTUs], Shannon index values of 5.21–6.16) and was significantly higher than that of Archaea (five phyla and 24 genera comprising 1141 OTUs and Shannon index values of 1.18–3.28). The bacterial communities were dominated by Proteobacteria (48.42–59.97% relative abundances), followed by Bacteroidetes (12.5–32.51%), Acidobacteria (2.07–11.56%), Firmicutes (0.65–6.32%), Planctomycetes (0.99–3.56%), Gemmatimonadetes (0.38–3.57%), Actinobacteria (1.67–3.52%), Verrucomicrobia (0.87–2.01%), and Chloroflexi (0.5–1.17%). In addition, archaeal communities were dominated by Thaumarchaeota (33.22–93.00%), followed by Euryarchaeota (2.89–35.47%), Woesearchaeota (0.99–31.04%), and Pacearchaeota (0.01–1.14%). The most abundant bacterial genus was Rhodoferax (5.73–26.62%) and the most abundant archaeal genus was the ammonia-oxidizing Nitrososphaera (29.18–91.46%). These results suggest that the Rhodoferax and Nitrososphaera are likely to participate in biogeochemical cycles in these environments through photoheterotrophy and nitrification, respectively. Taken together, these results provide valuable data for better understanding microbial interactions with each other and with these unique environments.




Similar content being viewed by others
References
Hayden CJ, Beman JM (2014) High abundances of potentially active ammonia-oxidizing bacteria and archaea in oligotrophic, high-altitude lakes of the Sierra Nevada, California, USA. PLoS ONE 9:e111560. https://doi.org/10.1371/journal.pone.0111560
Casamayor EO (2017) Towards a microbial conservation perspective in high mountain lakes. In: Catalan J, Ninot J, Aniz M (eds) High mountain conservation in a changing world. Advances in global change research vol 62. Springer, Cham, pp 157–180. https://doi.org/10.1007/978-3-319-55982-7_7
Cabrol NA, Mckay CP, Grin EA, Kiss KT, Ács E, Tóth B, Grigorszky I et al (2009) Signatures of habitats and life in earth’s high-altitude lakes: clues to Noachian aqueous environments on mars. In: Chapman MG (ed) The geology of mars: evidence from earth-based analogs. Cambridge University Press, UK, pp 350–370. https://doi.org/10.1017/CBO9780511536014
Yang Y, Shan J, Zhang J, Zhang X, Xie S, Liu Y (2014) Ammonia- and methane-oxidizing microorganisms in high-altitude wetland sediments and adjacent agricultural soils. Appl Microbiol Biotechnol 98:10197–10209. https://doi.org/10.1007/s00253-014-5942-x
Yun J, Ju Y, Deng Y, Zhang H (2014) Bacterial community structure in two permafrost wetlands on the Tibetan Plateau and Sanjiang Plain, China. Microb Ecol 68:360–369. https://doi.org/10.1007/s00248-014-0415-4
Aanderud ZT, Vert JC, Lennon JT, Lennon JT, Magnusson TW, Breakwell DP, Harker AR (2016) Bacterial dormancy is more prevalent in freshwater than hypersaline lakes. Front Microbiol 7:853. https://doi.org/10.3389/fmicb.2016.00853
Aszalós JM, Krett G, Anda D, Márialigeti K, Nagy B, Borsodi AK (2016) Diversity of extremophilic bacteria in the sediment of high-altitude lakes located in the mountain desert of Ojos del Salado volcano, dry-Andes. Extremophiles 20:603–620. https://doi.org/10.1007/s00792-016-0849-3
Hu A, Yao T, Jiao N, Liu Y, Yang Z, Liu X (2010) Community structures of ammonia-oxidising archaea and bacteria in high-altitude lakes on the Tibetan Plateau. Freshwater Biol 55:2375–2390. https://doi.org/10.1111/j.1365-2427.2010.02454.x
Dorador C, Vila I, Witzel KP, Imhoff JF (2013) Bacterial and archaeal diversity in high altitude wetlands of the Chilean Altiplano. Fundam Appl Limnol 182:135–159. https://doi.org/10.1127/1863-9135/2013/0393
Zhang J, Yang Y, Zhao L, Li Y, Xie S, Liu Y (2015) Distribution of sediment bacterial and archaeal communities in plateau freshwater lakes. Appl Microbiol Biotechnol 99:3291–3302. https://doi.org/10.1007/s00253-014-6262-x
Deng Y, Liu Y, Dumont M, Conrad R, Liu Y (2017) Salinity affects the composition of the aerobic methanotroph community in alkaline lake sediments from the Tibetan Plateau. Microb Ecol 73:101–110. https://doi.org/10.1007/s00248-016-0879-5
Zheng M, Liu X (2009) Hydrochemistry of salt lakes of the Qinghai-Tibet Plateau, China. Aquat Geochem 15:293–320. https://doi.org/10.1007/s10498-008-9055-y
Zhe M, Zhang X, Wang B, Sun R, Zheng D (2017) Hydrochemical regime and its mechanism in Yamzhog Yumco Basin, South Tibet. J Geogr Sci 27:1111–1122. https://doi.org/10.1007/s11442-017-1425-1
Xiong J, Liu Y, Lin X, Zhang H, Zeng J, Hou J, Yang Y, Yao T, Knight R, Chu H (2012) Geographic distance and pH drive bacterial distribution in alkaline lake sediments across Tibetan Plateau. Environ Microbiol 14:2457–2466. https://doi.org/10.1111/j.1462-2920.2012.02799.x
Liu Y, Yao T, Jiao N, Zhu LP, Hu AY, Liu XB, Gao J, Chen ZQ (2013) Salinity impact on bacterial community composition in five high-altitude lakes from the Tibetan Plateau, western China. Geomicrob J 30:462–469. https://doi.org/10.1080/01490451.2012.710709
Günther F, Thiele A, Gleixner G, Xu B, Yao T, Stefan S (2014) Distribution of bacterial and archaeal ether lipids in soils and surface sediments of Tibetan lakes: implications for GDGT-based proxies in saline high mountain lakes. Org Geochem 67:19–30. https://doi.org/10.1016/j.orggeochem.2013.11.014
Liu Y, Priscu JC, Xiong J, Conrad R, Hou J (2016) Salinity drives archaeal distribution patterns in high altitude lake sediments on the Tibetan Plateau. FEMS Microbiol Ecol 92:fiw033. https://doi.org/10.1093/femsec/fiw033
Yang J, Ma L, Jiang H, Wu G, Dong H (2016) Salinity shapes microbial diversity and community structure in surface sediments of the Qinghai-Tibetan lakes. Sci Rep 6:25078. https://doi.org/10.1038/srep25078
Zhong ZP, Liu Y, Miao LL, Wang F, Liu ZP (2016) Prokaryotic community structure driven by salinity and ionic concentrations in plateau lakes of the Tibetan Plateau. Appl Environ Microbiol 82:1846–1858. https://doi.org/10.1128/AEM.03332-15
Jiang H, Dong H, Yu B, Liu X, Li Y, Ji S, Zhang CL (2007) Microbial response to salinity change in Lake Chaka, a hypersaline lake on Tibetan Plateau. Environ Microbiol 9:2603–2621. https://doi.org/10.1111/j.1462-2920.2007.01377.x
Jiang H, Dong H, Deng S, Yu B, Huang Q, Wu Q (2009) Response of archaeal community structure to environmental changes in lakes on the Tibetan Plateau, northwestern China. Geomicrob J 26:289–297. https://doi.org/10.1080/01490450902892662
Liu X, Yao T, Kang S, Jiao N, Zeng Y, Liu Y (2010) Bacterial community of the largest oligosaline lake, Namco on the Tibetan Plateau. Geomicrob J 27:669–682. https://doi.org/10.1080/01490450903528000
Yang J, Jiang H, Dong H, Wang H, Wu G, Hou W, Liu W, Zhang C, Sun Y, Lai Z (2013) amoA-encoding archaea and thaumarchaeol in the lakes on the northeastern Qinghai-Tibetan Plateau. China Front Microbiol 4:329. https://doi.org/10.3389/fmicb.2013.00329
Liu Y, Priscu JC, Yao T, Vick-Majors TJ, Michaud AB, Jiao N, Hou J, Tian L, Hu A, Chen ZQ (2014) A comparison of pelagic, littoral, and riverine bacterial assemblages in lake Bangongco, Tibetan Plateau. FEMS Microbiol Ecol 89:211–221. https://doi.org/10.1111/1574-6941.12278
Liu X, Hou W, Dong H, Wang S, Jiang H, Wu G, Yang J, Li G (2015) Distribution and diversity of Cyanobacteria and eukaryotic algae in Qinghai-Tibetan lakes. Geomicrob J 33:860–869. https://doi.org/10.1080/01490451.2015.1120368
Han R, Zhang X, Liu J, Long Q, Chen L, Liu D, Zhu D (2017) Microbial community structure and diversity within hypersaline Keke salt lake environments. Can J Microbiol 63:895–908. https://doi.org/10.1139/cjm-2016-0773
Liu K, Liu Y, Jiao N, Zhu L, Wang J, Hu A, Liu X (2016) Vertical variation of bacterial community in Nam Co, a large stratified lake in central Tibetan Plateau. Antonie Van Leeuwenhoek 109:1323–1335. https://doi.org/10.1007/s10482-016-0731-4
Mitamura O, Seike Y, Kondo K, Goto N, Anbutsu K, Akatsuka T, Kihira M, Tsering TQ, Nishimura M (2003) First investigation of ultraoligotrophic alpine lake Puma Yum Co in the pre-Himalayas, China. Limnology 4:167–175. https://doi.org/10.1007/s10201-003-0101-6
Murakami T, Terai H, Yoshiyama Y, Tezuka T, Zhu L, Matsunaka T, Nishimura M (2007) The second investigation of lake Puma Yum Co located in the southern Tibetan Plateau, China. Limnol 8:331–335. https://doi.org/10.1007/s10201-007-0208-2
Liu Y, Yao T, Zhu L, Jiao NZ, Liu XB, Zeng YH, Jiang HC (2009) Bacterial diversity of freshwater alpine Lake Puma Yumco on the Tibetan Plateau. Geomicrob J 26:131–145. https://doi.org/10.1080/01490450802660201
Watanabe T, Matsunaka T, Nakamura T, Nishimura M, Izutsu Y, Minami M et al (2010) Last glacial-holocene geochronology of sediment cores from a high-altitude Tibetan lake based on AMS 14C dating of plant fossils: implications for paleoenvironmental reconstructions. Chem Geol 277:21–29. https://doi.org/10.1016/j.chemgeo.2010.07.004
Deng Y, Zhang Y, Gao Y, Li D, Liu R, Liu M, Zhang H, Hu B, Yu T, Yang M (2012) Microbial community compositional analysis for series reactors treating high level antibiotic wastewater. Environ Sci Technol 46:795–801. https://doi.org/10.1021/es2025998
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. https://doi.org/10.14806/ej.17.1.200
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/AEM.00062-07
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. https://doi.org/10.1128/AEM.01541-09
Asnicar F, Weingart G, Tickle TL, Huttenhower C, Segata N (2015) Compact graphical representation of phylogenetic data and metadata with GraPhlAn. Peer J 3:e1029. https://doi.org/10.7717/peerj.1029
Letunic I, Bork P (2015) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–245. https://doi.org/10.1093/nar/gkw290
Newton RJ, Jones SE, Eiler A, Mcmahon KD, Bertilsson S (2011) A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev 75:14–49. https://doi.org/10.1128/MMBR.00028-10
Auguet JC, Triadó-Margarit X, Nomokonova N, Camarero L, Casamayor EO (2012) Vertical segregation and phylogenetic characterization of ammonia-oxidizing archaea in a deep oligotrophic lake. ISME J 6:1786–1797. https://doi.org/10.1038/ismej.2012.33
Cao H, Auguet JC, Gu JD (2013) Global ecological pattern of ammonia-oxidizing archaea. PLoS ONE 8:e52853. https://doi.org/10.1371/journal.pone.0052853
Liu Y, Zhang J, Zhang X, Xie S (2014) Depth-related changes of sediment ammonia-oxidizing microorganisms in a high-altitude freshwater wetland. Appl Microbiol Biotechnol 98:5697–5707. https://doi.org/10.1007/s00253-014-5651-5
Liu Y, Zhang J, Zhao L, Li Y, Dai Y, Xie S (2015) Distribution of sediment ammonia-oxidizing microorganisms in plateau freshwater lakes. Appl Microbiol Biotechnol 99:4435–4444. https://doi.org/10.1007/s00253-014-6341-z
Hiraishi A, Imhoff JF (2015) Rhodoferax. In: Whitman WB, DeVos P, Dedysh S, et al. (eds) Bergey’s manual of systematics of archaea and bacteria. Wiley, Hoboken, pp 1–11. https://doi.org/10.1002/9781118960608.gbm00951
Salka I, Cuperová Z, Mašín M, Koblížek M, Grossart HP (2011) Rhodoferax-related pufM gene cluster dominates the aerobic anoxygenic phototrophic communities in German freshwater lakes. Environ Microbiol 13:2865–2875. https://doi.org/10.1111/j.1462-2920.2011.02562.x
Chong SC, Boone DR (2015) Methanoculleus. In: Whitman WB, DeVos P, Dedysh S, et al. (eds) Bergey’s manual of systematics of archaea and bacteria. Wiley, Hoboken, pp 1–5. https://doi.org/10.1002/9781118960608.gbm00505
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (31760034, 31860030, and 21967018), the Key Research Foundation of Development and Transformation of Qinghai Province (2019SF121), the Applied Basic Research Program of Qinghai Province (2018ZJ778, 2018ZJ930Q, and 2020ZJ767), and the Team’s Research Program of Microbial Resources in Salt-lakes of Qinghai-Tibetan Plateau (2018KYT1). We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research Involving Human Participants and/or Animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, R., Han, R., Long, Q. et al. Bacterial and Archaeal Communities within an Ultraoligotrophic, High-altitude Lake in the Pre-Himalayas of the Qinghai-Tibet Plateau. Indian J Microbiol 60, 363–373 (2020). https://doi.org/10.1007/s12088-020-00881-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-020-00881-8