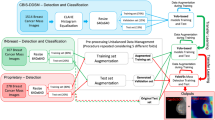
Abstract
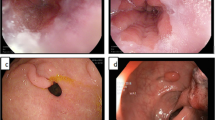
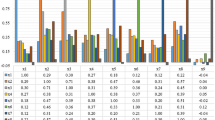
Computer aided diagnosis of cancer is a field of substantial worth in current scenario since approximately 38% population of the world is suffering from the disease. The detection of cancer is based on the observation of deformation in nuclei structure using histopathology slides/images. The proposed technique utilizes nuclei localization prior to classification of histopathology images as benign and malignant. The features used for classification are an ensemble of 150 bag of visual word features, extracted from preprocessed image and 20 handcrafted features, extracted from the internal parts of nuclei using localized histopathology images. The simulation results confirm the superiority of proposed localization based cancer classification method as compared to existing methods of the domain. It has reported average classification accuracy of 95.03% on BreakHis dataset.




Similar content being viewed by others
References
Jung, C., Kim, C.: Segmenting clustered nuclei using H-minima transform-based marker extraction and contour parameterization. IEEE Trans. Biomed. Eng. 57(10), 2600–2604 (2010)
Wang, P., Hu, X., Li, Y., Liu, Q., Zhu, X.: Automatic cell nuclei segmentation and classification of breast cancer histopathology images. Signal Process. 122, 1–13 (2016)
Wang, Z.: A semi-automatic method for robust and efficient identification of neighboring muscle cells. Pattern Recogn. 53, 300–312 (2016)
Jabeen, A., Riaz, M.M., Iltaf, N., Ghafoor, A.: Image contrast enhancement using weighted transformation function. IEEE Sens. J. 16(20), 7534–7536 (2016)
Nguyen, K., Sarkar, A., Jain, A.K.: Prostate cancer grading: use of graph cut and spatial arrangement of nuclei. IEEE Trans. Med. Imag 33(12), 2254–2270 (2014)
Lu, Z., Carneiro, G., Bradley, A.P., et al.: Evaluation of three algorithms for the segmentation of overlapping cervical cells. IEEE J. Biomed. Health Inf. 21(2), 441–450 (2017)
Spanhol, F.A., Oliveira, L.S., Petitjean, C., Heutte, L.: A dataset for breast cancer histopathological image classification. IEEE Trans. Biomed. Eng. 63(7), 1455–1462 (2016)
Vu, T.H., Mousavi, H.S., et al.: Histopathological image classification using discriminative feature-oriented dictionary learning. IEEE Trans. Med Imag 35(3), 738–751 (2016)
Naylor, P., La, M., Reyal, F., Walter, T.: Segmentation of nuclei in histopathology images by deep regression of the distance map. IEEE Trans. Med. Imag (2018). https://doi.org/10.21227/H26X0H
Li, X., Plataniotis, K.N.: Circular mixture modeling of color distribution for blind stain separation in pathology images. IEEE J. Biomed. Health Inf. 21(1), 150–161 (2017)
Wang, W., Ozolek, J.A., Slepcev, D., et al.: An optimal transportation approach for nuclear structure-based pathology. IEEE Trans. Med. Imag. 30(3), 621–631 (2011)
Dundar, M.M., Badve, S., Bilgin, G., et al.: Computerized classification of intraductal breast lesions using histopathological images. IEEE Trans. Biomed. Eng. 58(7), 1977–1984 (2011)
Basavanhally, A.N., et al.: Computerized image-based detection and grading of lymphocytic infiltration in HER2 + breast cancer histopathology. IEEE Trans. Biomed. Eng. 57(3), 642–653 (2010)
Manivannan, S., Li, W., Zhang, J., et al.: Structure prediction for gland segmentation with hand-crafted and deep convolutional features. IEEE Trans. Med. Imag 37(1), 210–221 (2018)
Cortes, C., Vapnik, V.: Support-vector networks. Mach. Learn. 20(3), 273–297 (1995)
Raji, C.G., Chandra, S.S.V.: Long-term forecasting the survival in liver transplantation using multilayer perceptron networks. IEEE Trans. Syst. Man Cybern. Syst. 47(8), 2318–2329 (2017)
Li, C., Wang, X., Liu, W., et al.: Weakly supervised mitosis detection in breast histopathology images using concentric loss. Med. Image Anal. 53, 165–178 (2019)
Yan, R., Ren, F., Wang, Z., et al.: Breast cancer histopathological image classification using a hybrid deep neural network. Methods (2019)
Yang, W., Wang, K., Zuo, W.: Neighborhood component feature selection for high-dimensional data. JCP 7, 161–168 (2012)
Klein, A., et al.: Fast Bayesian optimization of machine learning hyperparameters on large datasets, 2016. ArXiv:abs/1605.07079
Irshad, H., et al.: Methods for nuclei detection, segmentation, and classification in digital histopathology: a review current status and future potential. IEEE Rev. Biomed. Eng. 7, 97–114 (2014)
Azzopardi, G., Petkov, N.: Contour detection by CORF operator. In: ANN and Machine Learning ICANN 2012, Lecture Notes in Computer Science, vol. 7552, pp. 395–402. Springer, Heidelberg (2012)
Kurmi, Y., Chaurasia, V., Ganesh, N.: Tumor malignancy detection using histopathology imaging. J. Med. Imaging Radiat. Sci. (2019). https://doi.org/10.1016/j.jmir.2019.07.004
E. Mercan, S. Aksoy, L. G. Shapiro, et al.: Localization of diagnostically relevant regions of interest in whole slide images. In: 22nd International Conference on Pattern Recognition, Stockholm, pp. 1179–1184 (2014)
Riddle, N.C., et al.: Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 21(2), 147–163 (2011)
Kvilekval, K., Fedorov, D., Obara, B., et al.: Bisque: a platform for bioimage analysis and management. Bioinformatics 26(4), 544–552 (2010)
Kurmi, Y., Chaurasia, V.: Multifeature-based medical image segmentation. IET Image Proc. 12(8), 1491–1498 (2018)
Chaurasia, V., Chaurasia, V.: Statistical feature extraction based fast fractal image compression. J. Vis. Commun. Image Represent. 41, 87–95 (2016)
Jaccard, P.: The distribution of the flora in the alpine zone. New Phytol. 11, 37–50 (1912)
Taha, A.A., Hanbury, A.: An efficient algorithm for calculating the exact Hausdorff distance. IEEE Trans. Pattern Anal. Mach. Intell. 37(11), 2153–2163 (2015)
Fawcett, T.: An introduction to ROC analysis. Pattern Recog. Lett. 27, 861–874 (2006)
Acknowledgements
Autors are thankful to the supporting team of Jawaharlal Nehru Cancer Hospital & Research Center, (JNCH&RC) Bhopal, India. Specially Smt. Asha Joshi (Chairman), Smt. Divya Parashar (CEO & Research Coordinator), Dr. K. V. Pandya (Director), Dr. Pradeep Kolekar (Medical Director) and Mr. Rakesh Joshi (Additional Director), JNCH&RC Bhopal, India, for facilitating to work with patient data for dataset preparation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kurmi, Y., Chaurasia, V., Ganesh, N. et al. Microscopic images classification for cancer diagnosis. SIViP 14, 665–673 (2020). https://doi.org/10.1007/s11760-019-01584-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11760-019-01584-4