Abstract
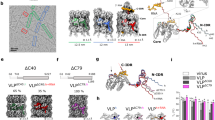
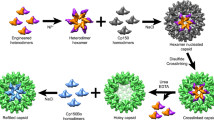
A variety of chemical approaches for the rational design of artificial proteins and peptides have been developed in recent years for the construction of self-assembled nanocapsules. It was previously found that a synthetic 24-mer β-annulus peptide, which participates in the formation of the dodecahedral internal skeleton of the tomato bushy stunt virus capsid, spontaneously self-assembled into artificial viral capsids with a size of 30–50 nm. These artificial viral capsids were established to encapsulate various guest molecules, such as anionic dyes, DNA, quantum dots, and His-tagged proteins. The artificial viral capsids could also be dressed up with gold nanoparticles, single-stranded DNA, coiled-coil spikes, and proteins by modifying with these molecules at the C-terminus of β-annulus peptides. The artificial viral capsids were notably stabilized by dressing up with human serum albumin and acquired enzymatic activity by dressing up with ribonuclease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Manchester M, Steinmetz NF, editors. Viruses and nanotechnology: current topics in microbiology and immunology. Belrin: Springer; 2009.
Khudyakov Y, Pumpens P, editors. Viral nanotechnology. Boca Raton, Florida: CRC Press; 2016.
Douglas T, Young M. Viruses: making friends with old foes. Science. 2006;312:873–5.
Steinmetz NF, Evans DJ. Utilisation of plant viruses in bionanotechnology. Org Biomol Chem. 2007;5:2891–902.
Papapostolou D, Howorka S. Engineering and exploiting protein assemblies in synthetic biology. Mol BioSyst. 2009;5:723–32.
Witus LS, Francis MB. Using synthetically modified proteins to make new materials. Acc Chem Res. 2011;44:774–83.
Bronstein LM. Virus-based nanoparticles with inorganic cargo: what does the future hold? Small. 2011;7:1609–18.
van Rijn P, Schirhagl R. Viruses, artificial viruses and virus-based structures for biomedical applications. Adv Healthc Mater. 2016;5:1386–400.
Azuma Y, Edwardson TGW, Hilvert D. Tailoring lumazine synthase assemblies for bionanotechnology. Chem Soc Rev. 2018;47:3543–57.
Douglas T, Young M. Host–guest encapsulation of materials by assembled virus protein cages. Nature. 1998;393:152–5.
Aragonès MC, Engelkamp H, Claessen VI, Sommerdijik NAJM, Rowan AE, Christianen PCM, Maan JC, Verduin BJM, Cornelissen JJLM, Nolte RJM. A virus-based single-enzyme nanoreactor. Nat Nanotechnol. 2007;2:635–9.
Matsuura K. Synthetic approaches to construct viral capsid-like spherical nanomaterials. Chem Commun. 2018;54:8944–59.
Matsuurua K. Rational design of self-assembled proteins and peptides for nano- and micro-sized architectures. RSC Adv. 2014;4:2942–53.
Luo Q, Hou C, Bai Y, Wang R, Liu J. Protein assembly: versatile approaches to construct highly ordered nanostructures. Chem Rev. 2016;116:13571–632.
Kuan SL, Bergamini FRG, Weil T. Functional protein nanostructures: a chemical toolbox. Chem Soc Rev. 2018;47:9069–105.
Sainsbury F. Emergence by design in self-assembling protein shells. ACS Nano. 2020;14:2565–8.
Lai YT, Tsai KL, Sawaya MR, Asturias FJ, Yeates TO. Structure and flexibility of nanoscale protein cages designed by symmetric self-assembly. J Am Chem Soc. 2013;135:7738–43.
Lai YT, Reading E, Hura GL, Tsai KL, Laganowsky A, Asturias FJ, Tainer JA, Robinson CV, Yeates TO. Structure of a designed protein cage that self-assembles into a highly porous cube. Nat Chem. 2014;6:1065–71.
Hsia Y, Bale JB, Gonen S, Shi D, Sheffler W, Fong KK, Nattermann U, Xu C, Huang PS, Ravichandran R, Yi S, Davis TN, Gonen T, King NP, Baker D. Design of a hyperstable 60-subunit protein icosahedron. Nature. 2016;535:136–9.
Kawakami N, Kondo H, Matsuzawa Y, Hayasaka K, Nasu E, Sasahara K, Arai R, Miyamoto K. Design of hollow protein nanoparticles with modifiable interior and exterior surfaces. Angew Chem Int Ed Engl. 2018;57:12400–4.
Cristie-David AS, Chen J, Nowak DB, Bondy AL, Sun K, Park SI, Holl MMB, Su M, Marsh ENG. Coiled-coil-mediated assembly of an icosahedral protein cage with extremely high thermal and chemical stability. J Am Chem Soc. 2019;141:9207–16.
Malay AD, Miyazaki N, Biela A, Chakraborti S, Majsterkiewicz K, Stupka I, Kaplan CS, Kowalczyk A, Piette BMAG, Hochberg GKA, Wu D, Wrobel TP, Fineberg A, Kushwah MS, Kelemen M, Vavpetič P, Pelicon P, Kukura P, Benesch JLP, Iwasaki K, Heddle JG. An ultra-stable gold-coordinated protein cage displaying reversible assembly. Nature. 2019;569:438–42.
Ramakers BEI, van Hest JCM, Löwik DWPM. Molecular tools for the construction of peptide-based materials. Chem Soc Rev. 2014;43:2743–56.
De Santis E, Ryadnov MG. Peptide self-assembly for nanomaterials: the old new kid on the block. Chem Soc Rev. 2015;44:8288–300.
Lou S, Wang X, Yu Z, Shi L. Peptide tectonics: encoded structural complementarity dictates programmable self-assembly. Adv Sci. 2019;6:1802043.
Fletcher JM, Harniman RL, Barnes FRH, Boyle AL, Collins A, Mantell J, Sharp TH, Antognozzi M, Booth PJ, Linden N, Miles MJ, Sessions RB, Verkade P, Woolfson DN. Self-assembling cages from coiled-coil peptide modules. Science. 2013;340:595–9.
Ross JF, Bridges A, Fletcher JM, Shoemark D, Alibhai D, Bray HEV, Beesley JL, Dawson WM, Hodgson LR, Mantell J, Verkade P, Edge CM, Sessions RB, Tew D, Woolfson DN. Decorating self-assembled peptide cages with proteins. ACS Nano. 2017;11:7901–14.
De Santis E, Alkassem H, Lamarre B, Faruqui N, Bella A, Noble JE, Micale N, Ray S, Burns JR, Yon AR, Hoogenboom BW, Ryadnov MG. Antimicrobial peptide capsids of de novo design. Nat Commun. 2017;8:2263.
Kepiro IE, Marzuoli I, Hammond K, Ba X, Lewis H, Shaw M, Gunnoo SB, De Santis E, Łapińska U, Pagliara S, Holmes MA, Lorenz CD, Hoogenboom BW, Fraternali F, Ryadnov MG. Engineering chirally blind protein pseudocapsids into antibacterial persisters. ACS Nano. 2020;14:1609–22.
Matsuura K, Murasato K, Kimizuka N. Artificial peptide-nanospheres self-assembled from three-way junctions of β-sheet-forming peptides. J Am Chem Soc. 2005;127:10148–9.
Matsuura K, Watanabe K, Sakurai K, Matsuzaki T, Kimizuka N. Self-assembled synthetic viral capsids from a 24-mer viral peptide fragment. Angew Chem Int Ed Engl. 2010;49:9662–5.
Olson AJ, Bricogne G, Harrison SC. Structure of tomato busy stunt virus IV. The virus particle at 2.9 A resolution. J Mol Biol. 1983;171:61–93.
Hopper P, Harrison SC, Sauer RT. Structure of tomato bushy stunt virus. V. Coat protein sequence determination and its structural implications. J Mol Biol. 1984;177:701–13.
Matsuura K, Watanabe K, Matsushita Y, Kimizuka N. Guest-binding behavior of peptide nanocapsules self-assembled from viral peptide fragments. Polym J. 2013;45:529–34.
Fujita S, Matsuura K. Encapsulation of CdTe quantum dots into synthetic viral capsids. Chem Lett. 2016;45:922–4.
Fujita S, Matsuura K. Inclusion of zinc oxide nanoparticles into virus-like peptide nanocapsules self-assembled from viral β-annulus peptide. Nanomaterials. 2017;4:778–91.
Matsuura K, Nakamura T, Watanabe K, Noguchi T, Minamihata K, Kamiya N, Kimizuka N. Self-assembly of Ni-NTA-modified β-annulus peptides into artificial viral capsids and encapsulation of His-tagged proteins. Org Biomol Chem. 2016;14:7869–74.
Nakamura Y, Inaba H, Matsuura K. Construction of artificial viral capsids encapsulating short DNAs via disulfide bonds and controlled release of DNAs by reduction. Chem Lett. 2019;48:544–6.
Matsuura K, Ueno G, Fujita S. Self-assembled artificial viral capsid decorated with gold nanoparticles. Polym J. 2015;47:146–51.
Fujita S, Matsuura K. Self-assembled artificial viral capsids bearing coiled-coils at the surface. Org Biomol Chem. 2017;15:5070–7.
Nakamura Y, Yamada S, Nishikawa S, Matsuura K. DNA-modified artificial viral capsids self-assembled from DNA-conjugated β-annulus peptide. J Pept Sci. 2017;23:636–43.
Matsuura K, Honjo T. Artificial viral capsid dressed up with human serum albumin. Bioconj Chem. 2019;30:1636–41.
Raines RT, Ribonuclease A, Chem Rev. 1998;98:1045–66.
Matsuura K, Ota J, Fujita S, Shiomi Y, Inaba H. Construction of ribonuclease-decorated artificial virus-like capsid by peptide self-assembly. J Org Chem. 2020;85:1668–73.
Acknowledgements
This research was partially supported by The Asahi Glass Foundation, a Grant-in-Aid for Scientific Research on Innovative Areas “Chemistry for Multimolecular Crowding Biosystems” (JSPS KAKENHI Grant No. JP18H04558), and Grant-in-Aid for Scientific Research (B) (JSPS KAKENHI Grant No. JP18H02089). I would like to thank Prof. Nobuo Kimizuka (Kyushu University), Dr Hiroshi Inaba (Tottori University) and all the coworkers, whose names are cited in the references, for their valuable contributions. The author would like to thank MARUZEN-YUSHODO Co., Ltd, for English language editing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares that he has no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Matsuura, K. Dressing up artificial viral capsids self-assembled from C-terminal-modified β-annulus peptides. Polym J 52, 1035–1041 (2020). https://doi.org/10.1038/s41428-020-0355-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41428-020-0355-4