Abstract
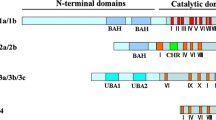
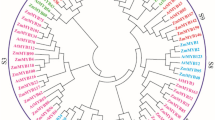
Methyl-CpG-binding domain (MBD) proteins in plants are important trans-acting factors specifically recognizing methylated DNA. The MBD proteins can compact chromatins to repress transcription by recruiting chromatin-modifying complexes that contain histone deacetylase activities and chromatin remodeling factors, and play a crucial biological role in the growth and development in plants. Currently, very little is known regarding the structure and function of MBD genes in plants. In this study, we performed a genome-wide identification and expression profile analysis of maize MBD genes (ZmMBDs) from the latest version of the maize (B73) genome. By analyzing phylogenetic relationship of MBD gene families from Arabidopsis, rice, wheat, and maize, all 14 MBD proteins in maize were categorized into four subclasses. Furthermore, chromosome location and schematic structure revealed an unevenly distribution on chromosomes and structure features of MBD genes in maize, respectively. Eventually, EST expression data mining, microarray data clustering analysis, and semi-quantitative and quantitative expression profile analyses detected in seedling leaves and stems by heat, drought, and salt-stress treatments have demonstrated that these genes had temporal and spatial expression pattern and exhibited different expression levels under heat, drought, and salt-stress conditions, suggesting that functional diversification of the MBD gene family in maize. In addition, through electrophoretic mobility shift assay (EMSA) a representative MBD protein, ZmMBD11, exhibited in vitro DNA-binding activity, indicating that that the MBD proteins in maize might play a role in reading cytosine methylation. Taken together, these results would provide an important theoretical basis for future functional verification of ZmMBD genes and also facilitate future experimental research to further unravel the mechanisms of epigenetic regulation in plants.










Similar content being viewed by others
References
Ballestar E, Wolffe AP (2001) Methyl-CpG-binding proteins. Targeting specific gene repression. Eur J Biochem 268:1–6
Berg A, Meza TJ, Mahić M, Thorstensen T, Kristiansen K, Aalen RB (2003) Ten members of the Arabidopsis gene family encoding methyl-CpG-binding domain proteins are transcriptionally active and at least one, AtMBD11, is crucial for normal development. Nucleic Acids Res 31:5291–5304
Cai R, Zhang C, Zhao Y, Zhu K, Wang Y, Jiang H, Xiang Y, Cheng B (2016) Genome-wide analysis of the IQD gene family in maize. Mol Genet Genomics 291(2):543–558
Campi M, D'Andrea L, Emiliani J, Casati P (2012) Participation of chromatin-remodeling proteins in the repair of ultraviolet-B-damaged DNA. Plant Physiol 158(2):981–995
Casati P, Walbot V (2008) Maize lines expressing RNAi to chromatin remodeling factors are similarly hypersensitive to UV-B radiation but exhibit distinct transcriptome responses. Epigenetics 3(4):216–229
Casati P, Stapleton AE, Blum JE, Walbot V (2006) Genome-wide analysis of high-altitude maize and gene knockdown stocks implicates chromatin remodeling proteins in response to UV-B. Plant J 46(4):613–627
Casati P, Campi M, Chu F, Suzuki N, Maltby D, Guan S, Burlingame AL, Walbot V (2008) Histone acetylation and chromatin remodeling are required for UV-B-dependent transcriptional activation of regulated genes in maize. Plant Cell 20(4):827–842
Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A (2006) Pfam: clans, web tools and services. Nucleic Acids Res 34(Database issue): D247–251
Finnegan EJ, Peacock WJ, Dennis ES (1996) Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci USA 93(16):8449–8454
Finnegan EJ, Genger RK, Peacock WJ, Dennis ES (1998) DNA methylation in plants. Annu Rev Plant Physiol Plant Mol Biol 49(1):223–247
Grafi G, Zemach A, Pitto L (2007) Methyl-CpG-binding domain (MBD) proteins in plants. Biochim Biophys Acta 1769(5–6):287–294
Gu Z, Cavalcanti A, Chen FC, Bouman P, Li WH (2002) Extent of gene duplication in the genomes of drosophila, nematode, and yeast. Mol Biol Evol 19:256–262
Ito M, Koike A, Koizumi N, Sano H (2003) Methylated DNA-binding proteins from Arabidopsis. Plant Physiol 133(4):1747–1754
Juretic N, Hoen DR, Huynh ML, Harrison PM, Bureau TE (2005) The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res 15(9):1292–1297
Kidwell MG, Lisch D (2001) Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution 55(1):1–24
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5(2):150–163
Lang Z, Lei M, Wang X, Tang K, Miki D, Zhang H, Mangrauthia SK, Liu W, Nie W, Ma G, Yan J, Duan CG, Hsu CC, Wang C, Tao WA, Gong Z, Zhu JK (2015) The methyl-CpG-binding protein MBD7 facilitates active DNA demethylation to limit DNA hyper-methylation and transcriptional gene silencing. Mol Cell 57(6):971–983
Le Guezennec X, Vermeulen M, Brinkman AB, Hoeijmakers WA, Cohen A, Lasonder E, Stunnenberg HG (2006) MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol 26(3):843–851
Letunic I, Doerks T, Bork P (2009) SMART 6: recent updates and new developments. Nucleic Acids Res 37(Database issue): D229–D232
Li Y, Meng F, Yin J, Liu H, Si Z, Ni Z, Sun Q, Ren J, Niu H (2008) Isolation and comparative expression analysis of six MBD genes in wheat. Biochim Biophys Acta 1779(2):90–98
Li Q, Wang X, Sun H, Zeng J, Cao Z, Li Y, Qian W (2015) Regulation of Active DNA Demethylation by a Methyl-CpG-Binding Domain Protein in Arabidopsis thaliana. PLoS Genet 11(5):e1005210
Li D, Palanca AMS, Won SY, Gao L, Feng Y, Vashisht AA, Liu L, Zhao Y, Liu X, Wu X, Li S, Le B, Kim YJ, Yang G, Li S, Liu J, Wohlschlegel JA, Guo H, Mo B, Chen X, Law JA (2017) The MBD7 complex promotes expression of methylated transgenes without significantly altering their methylation status. Elife 6:e19893
Lippman Z, May B, Yordan C, Singer T, Martienssen R (2003) Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PloS Genet 1:420–428
May BP, Lippman ZB, Fang Y, Spector DL, Martienssen RA (2005) Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet 1(6):e79
Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S (2004) Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol 24(21):9630–9645
Qian Y, Cheng Y, Cheng X, Jiang H, Zhu S, Cheng B (2011) Identification and characterization of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families in maize. Plant Cell Rep 30(7):1347–1363
Qian Y, Xi Y, Cheng B, Zhu S (2014a) Genome-wide identification and expression profiling of DNA methyltransferase gene family in maize. Plant Cell Rep 33(10):1661–1672
Qian Y, Xi Y, Cheng B, Zhu S, Kan X (2014b) Identification and characterization of the SET domain gene family in maize. Mol Biol Rep 41(3):1341–1354
Questa JI, Rius SP, Casadevall R, Casati P (2016) ZmMBD101 is a DNA-binding protein that maintains Mutator elements chromatin in a repressive state in maize. Plant Cell Environ 39(1):174–184
Scebba F, Bernacchia G, De Bastiani M, Evangelista M, Cantoni RM, Cella R, Locci MT, Pitto L (2003) Arabidopsis MBD proteins show different binding specificities and nuclear localization. Plant Mol Biol 53:755–771
Scebba F, De Bastiani M, Bernacchia G, Andreucci A, Galli A, Pitto L (2007) PRMT11: a new Arabidopsis MBD7 protein partner with arginine methyltransferase activity. Plant J 52(2):210–222
Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM (2011) Genome-wide atlas of transcription during maize development. Plant J 66(4):553–563
Soderlund C, Bomhoff M, Nelson WM (2011) SyMAP v34: a turnkey synteny system with application to plant genomes. Nucleic Acids Res 39(10):e68
Springer NM, Kaeppler SM (2005) Evolutionary divergence of monocot and dicot methyl-CpG-binding domain proteins. Plant Physiol 138(1):92–104
Thompson J, Higgins D, Gibson T (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Van Bel M, Proost S, Wischnitzki E, Movahedi S, Scheerlinck C, Van de Peer Y, Vandepoele K (2012) Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol 158(2):590–600
Wade PA, Wolffe AP (2001) Recognizing methylated DNA. Nat Struc Biol 8:575–577
Wang Y, Deng D, Bian Y, Lv Y, Xie Q (2010) Genome-wide analysis of primary auxin-responsive Aux/IAA gene family in maize (Zea mays L.). Mol Biol Rep 37:3991–4001
Wang C, Dong X, Jin D, Zhao Y, Xie S, Li X, He X, Lang Z, Lai J, Zhu JK, Gong Z (2015) Methyl-CpG-binding domain protein MBD7 is required for active DNA demethylation in Arabidopsis. Plant Physiol 167(3):905–914
Wei F, Coe E, Nelson W, Bharti AK, Engler F, Butler E, Kim H, Goicoechea JL, Chen M, Lee S, Fuks G, Sanchez-Villeda H, Schroeder S, Fang Z, McMullen M, Davis G, Bowers JE, Paterson AH, Schaeffer M, Gardiner J, Cone K, Messing J, Soderlund C, Wing RA (2007) Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet 3(7):e123
Yang S, Zhang X, Yue J, Tian D, Chen J (2008) Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Genet Genomics 280:187–198
Zemach A, Grafi G (2003) Characterization of Arabidopsis thaliana methyl-CpG-binding domain (MBD) proteins. Plant J 34:565–572
Zemach A, Grafi G (2007) Methyl-CpG-binding domain proteins in plants: interpreters of DNA methylation. Trends Plant Sci 12(2):80–85
Zemach A, Li Y, Wayburn B, Ben-Meir H, Kiss V, Avivi Y, Kalchenko V, Jacobsen SE, Grafi G (2005) DDM1 binds Arabidopsis methyl-CpG binding domain proteins and affects their subnuclear localization. Plant Cell 17(5):1549–1558
Zhang X, Feng Y, Cheng H, Tian D, Yang S, Chen J (2011) Relative evolutionary rates of NBS-encoding genes revealed by soybean segmental duplication. Mol Genet Genomics 285:79–90
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (No.31571673) and the Anhui Provincial University Excellent Youth Talent Support Program (No. 06146007) and the open funding of National Engineering Laboratory of Crop Stress Resistance Breeding (No.KNZJ1023). The authors are grateful to the reviewers and editors for their helpful comments on this paper.
Author information
Authors and Affiliations
Contributions
These studies were designed by Dr. QJ LY, CCL, RQY, ZJ, and CL carried out all the experimental analyses and prepared all figures and tables. The manuscript was drafted by Dr. Q. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Qian, Y., Ren, Q., Jiang, L. et al. Genome-wide analysis of maize MBD gene family and expression profiling under abiotic stress treatment at the seedling stage. Plant Biotechnol Rep 14, 323–338 (2020). https://doi.org/10.1007/s11816-020-00607-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-020-00607-8