Abstract
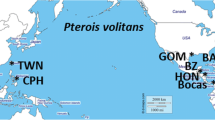
As documented marine invasions rise, determining the underlying molecular mechanisms of successful invasions is an immediate concern to management, but the relevance of such genetic studies hinges upon choosing the right markers. Most invasive lionfish phylogeography studies have used only the mitochondrial D-loop marker. For this study, three markers were targeted in invasive lionfish (n = 148): D-loop (mitochondrial), cytochrome oxidase I (mitochondrial), and S7 ribosomal intron-1 (nuclear). Using both mitochondrial and nuclear markers showed evidence of hybridization. At the D-loop marker, all sequences grouped with the Pacific Ocean lineage represented by Pterois volitans. At the S7 intron-1, all sequences grouped with the Indian Ocean S7 intron-1 haplotype associated with the Indian Ocean lineage represented by Pterois miles, showing discordance between the mitochondrial and nuclear markers. Further, at the COI marker, three out of 163 individuals sequenced matched P. miles. High throughput sequencing of the entire mitochondrial genome (n = 7) revealed that incongruence between mtDNA markers may be due to the insertion of mitochondrial DNA from P. volitans into the nuclear genome of fish genetically identified as P. miles. These data suggest hybridization and demonstrate that careful marker choice is important in future studies of lionfish genetics and invasive species in general.








Similar content being viewed by others
References
Aguilar-Perera A, Tuz-Sulub A (2010) Non-native, invasive red lionfish (Pterois volitans [Linnaeus, 1758]: Scorpaenidae), is first recorded in the southern Gulf of Mexico, off the northern Yucatan Peninsula, Mexico. Aquat Invasions 5:S9–S12
Albins MA, Hixon MA (2008) Invasive Indo-Pacific lionfish Pterois volitans reduce recruitment of Atlantic coral-reef fishes. Mar Ecol Prog Ser 367:233–238
Albins MA, Hixon MA (2013) Worst case scenario: potential long-term effects of invasive predatory lionfish (Pterois volitans) on Atlantic and Caribbean coral-reef communities. Environ Biol Fishes 96:1151–1157
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Antunes A, Ramos MJ (2005) Discovery of a large number of previously unrecognized mitochondrial pseudogenes in fish genomes. Genomics 86:708–717
Bax N, Williamson A, Aguero M, Gonzalez E, Geeves W (2003) Marine invasive alien species: a threat to global biodiversity. Mar Policy 27:313–323
Bensasson DD, Zhang DX, Hartl DL, Hewitt GM (2001) Mitochondrial pseudogenes: evolution’s misplaced witnesses. Trends Ecol Evol 16:314–321
Betancur-R R, Hines A, Acero P, Ortí G, Wilbur AE, Freshwater DW (2011) Reconstructing the lionfish invasion: insights into Greater Caribbean biogeography. J Biogeogr 38:1281–1293
Bock G, Caseys C, Cousens RD, Hahn MA, Heredia SM, Hübner S, Turner KG, Whitney KD, Rieseberg LH (2015) What we still don’t know about invasion genetics. Mol Ecol 24:2277–2297
Boros G, Mozsár A, Vitál Z, Nagy AS, Specziár A (2014) Growth and condition factor of hybrid (Bighead Hypophthalmichthys nobilis Richardson, 1845× silver carp H. molitrix Valenciennes, 1844) Asian carps in the shallow, oligo-mesotrophic Lake Balaton. J Appl Ichthyol 30:546–548
Bors EK, Herrera S, Morris JA Jr, Shank TM (2019) Population genomics of rapidly invading lionfish in the Caribbean reveals signals of range expansion in the absence of spatial population structure. Ecol Evolut. https://doi.org/10.1002/ece3.4952
Butterfield JSS, Díaz-Ferguson E, Silliman BR, Saunders JW, Buddo D, Mignucci-Giannoni AA, Searle L, Allen AC, Hunter ME (2015) Wide-ranging phylogeographic structure of invasive red lionfish in the Western Atlantic and Greater Caribbean. Mar Biol 162:773–781
Clement M, Posada DCKA, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Consuegra S, John E, Verspoor E, De Leaniz CG (2015) Patterns of natural selection acting on the mitochondrial genome of a locally adapted fish species. Genet Select Evolut 47:58
Côté IM, Maljković A (2010) Predation rates of Indo-Pacific lionfish on Bahamian coral reefs. Mar Ecol Prog Ser 404:219–225
Cristescu ME (2015) Genetic reconstructions of invasion history. Mol Ecol 24:2212–2225
Darling E, Green S, O’Leary JK, Côté I (2011) M. Indo-Pacific lionfish are larger and more abundant on invaded reefs: a comparison of Kenyan and Bahamian lionfish populations. Biol Invasions 13:2045–2051
Dudu A, Georgescu SE, Berrrebi P, Costache M (2012) Site heteroplasmy in the mitochondrial cytochrome b gene of the sterlet sturgeon Acipenser ruthenus. Genet Mol Biol 35:886–891
Ferreira CEL, Luiz OJ, Floeter SR, Lucena MB, Barbosa MC, Rocha CR, Rocha LA (2015) First record of invasive lionfish (Pterois volitans) for the Brazilian coast. PLoS ONE 10:e0123002
Freshwater DW, Hines A, Parham S, Wilbur AE, Sabaoun M, Woodhead J, Akins JL, Purdy B, Whitfield PE, Paris CB (2009a) Mitochondrial control region sequence analyses indicate dispersal from the US East Coast as the source of the invasive Indo-Pacific lionfish Pterois volitans in the Bahamas. Mar Biol 156:1213–1221
Freshwater DW, Hamner RM, Parham S, Wilbur AE (2009b) Molecular evidence that the lionfishes Pterois miles and Pterois volitans are distinct species. J North Carol Acad Sci 125:39–46
Galtier N, Nabholz B, Glémin S, Hurst GDD (2009) Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol 18:4541–4550
Green SJ, Akins JL, Maljkovic´ A, Coˆte´ IM, (2012) Invasive lionfish drive Atlantic coral reef fish declines. PLoS ONE 7:e32596
Green SJ, Akins JL, Côté IM (2011) Foraging behaviour and prey consumption in the Indo-Pacific lionfish on Bahamian coral reefs. Mar Ecol Prog Ser 433:159–167
Green SJ, Coˆte´ IM, (2008) Record densities of Indo-Pacific lionfish on Bahamian coral reefs. Coral Reefs 28:107
Good JM, Vanderpool D, Keeble S, Bi K (2015) Negligible nuclear introgression despite complete mitochondrial capture between two species of chipmunks. Evolution 69(8):1961–1972
Gurevitch J, Padilla DK (2004) Are invasive species a major cause of extinctions? Trends Ecol Evol 19:470–474
Hahn MA, Rieseberg LH (2017) Genetic admixture and heterosis may enhance the invasiveness of common ragweed. Evol Appl 10:241–250
Hamner RM, Freshwater DW, Whitfield PE (2007) Mitochondrial cytochrome b analysis reveals two invasive lionfish species with strong founder effects in the western Atlantic. J Fish Biol 71:214–222
Hazkani-Covo E, Covo S (2008) Numt-mediated double-strand break repair mitigates deletions during primate genome evolution. PLoS Genet 4(10):e1000237
Hazkani-Covo E, Zeller RM, Martin W (2010) Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet 6(2):e1000834
Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN (2007) Universal primer cocktails for fish DNA barcoding. Mol Ecol Notes 7:544–548
Johnson J, Bird C, Johnston MA, Fogg AQ, Hogan JD (2016) Regional genetic structure and genetic founder effects in the invasive lionfish: comparing the Gulf of Mexico. Caribb N Atl Mar Biol 163:216
Kochzius M, Söller R, Khalaf MA, Blohm D (2003) Molecular phylogeny of the lionfish genera Dendrochirus and Pterois (Scorpaenidae, Pteroinae) based on mitochondrial DNA sequences. Mol Phylogenet Evol 28:396–403
Kolbe JJ, Larson A, Losos JB, de Queiroz K (2008) Admixture determines genetic diversity and population differentiation in the biological invasion of a lizard species. Biol Lett 4:434–437
Lamer JT, Dolan CR, Petersen JL, Chick JH, Epifanio JM (2010) Introgressive hybridization between bighead carp and silver carp in the Mississippi and Illinois Rivers. N Am J Fish Manag 30:1452–1461
Layman CA, Jud ZR, Nichols P (2014) Lionfish alter benthic invertebrate assemblages in patch habitats of a subtropical estuary. Mar Biol 161:2179–2182
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Liang B, Wang N, Li N, Kimball RT, Braun EL (2018) Comparative Genomics Reveals a Burst of Homoplasy-Free Numt Insertions. Mol Biol Evol 35(8):2060–2064
Lindholm AK, Breden F, Alexander HJ, Chan W-K, Thakurta SG, Brooks R (2005) Invasion success and genetic diversity of introduced populations of guppies Poecilia reticulata in Australia. Mol Ecol 14:3671–3682
McCormick MI, Allan BJM (2016) Lionfish misidentification circumvents an optimized escape response by prey. Conserv Physiol 4:cow064
McDonald JH, Kreitman M (1991) Neutral mutation hypothesis test. Nature 354:6349
McGeoch MA, Butchart SHM, Spear D, Marais E, Kleynhans EJ, Symes A, Chanson J, Hoffmann M (2010) Global indicators of biological invasion: species numbers, biodiversity impact and policy responses. Divers Distrib 16:95–108
Molnar JL, Gamboa RL, Revenga C, Spalding MD (2008) Assessing the global threat of invasive species to marine biodiversity. Front Ecol Environ 6:485–492
Morgan JA, Macbeth M, Broderick D, Whatmore P, Street R, Welch DJ, Ovenden JR (2013) Hybridisation, paternal leakage and mitochondrial DNA linearization in three anomalous fish (Scombridae). Mitochondrion 13:852–861
Moritz CTED, Dowling TE, Brown WM (1987) Evolution of animal mitochondrial DNA: relevance for population biology and systematics. Annu Rev Ecol Syst 18:269–292
Morris JA, Akins JL, Barse A, Cerino D, Freshwater DW, Green SJ, Muñoz RC, Paris C, Whitfield PE (2009) Biology and ecology of the invasive lionfishes, Pterois miles and Pterois volitans. Proc Gulf Caribb Fish Inst 1(61):409–414
Mourier T, Hansen AJ, Willerslev E, Arctander P (2001) The Human Genome Project reveals a continuous transfer of large mitochondrial fragments to the nucleus. Mol Biol Evol 18(9):1833–1837
Nielsen HM, Ødegård J, Olesen I, Gjerde B, Ardo L, Jeney G, Jeney Z (2010) Genetic analysis of common carp (Cyprinus carpio) strains: I: genetic parameters and heterosis for growth traits and survival. Aquaculture 304:14–21
Pérez-Portela R, Bumford A, Coffman B, Wedelich S, Davenport M, Fogg A, Swenarton MK, Coleman F, Johnston MA, Crawford DL, Oleksiak MF (2018) Genetic homogeneity of the invasive lionfish across the Northwestern Atlantic and the Gulf of Mexico based on Single Nucleotide Polymorphisms. Sci Rep 8:5062
Ricchetti M, Tekaia F, Dujon B (2004) Continued colonization of the human genome by mitochondrial DNA. PLoS Biol 2(9):e273
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Ruttenberg BI, Schofield PJ, Akins JL, Acosta A, Feeley MW, Blondeau J, Smith SG, Ault JS (2012) Rapid invasion of Indo-Pacific lionfishes (Pterois volitans and Pterois miles) in the Florida Keys, USA: evidence from multiple pre-and post-invasion data sets. Bull Mar Sci 88:1051–1059
Schofield PJ (2009) Geographic extent and chronology of the invasion of non-native lionfish (Pterois volitans [Linnaeus 1758] and P. miles [Bennett 1828]) in the Western North Atlantic and Caribbean Sea. Aquat Invasions 4:473–0479
Schultz ET (1986) Pterois volitans and Pterois miles: two valid species. Copeia 3:686–690
Silva G, Lima FP, Martel P, Castilho R (2014) Thermal adaptation and clinal mitochondrial DNA variation of European anchovy. Proc R Soc Lond B 281:20141093
Smith KF, Abbott CL, Saito Y, Fidler AE (2015) Comparison of whole mitochondrial genome sequences from two clades of the invasive ascidian, Didemnum vexillum. Mar Genomics 19:75–83
Sutherland WJ, Clout M, Côté IM et al (2010) A horizon scan of global conservation issues for 2010. Trends Ecol Evol 25:1–7
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tang CQ, Leasi F, Obertegger U, Kieneke A, Barraclough TG, Fontaneto D (2012) The widely used small subunit 18S rDNA molecule greatly underestimates true diversity in biodiversity surveys of the meiofauna. Proc Natl Acad Sci USA 109:16208–16212
Teletchea F, Laudet V, Hänni C (2006) Phylogeny of the Gadidae (sensu Svetovidov, 1948) based on their morphology and two mitochondrial genes. Mol Phylogenet Evol 38:189–199
Toledo-Hernández C, Vélez-Zuazo X, Ruiz-Diaz CP, Patricio AR, Mège P, Navarro M, Sabat AM, Betancur-R R, Papa R (2014) Population ecology and genetics of the invasive lionfish in Puerto Rico. Aquat Invasions 9:227–237
Tourmen Y, Baris O, Dessen P, Jacques C, Malthièry Y, Reynier P (2002) Structure and chromosomal distribution of human mitochondrial pseudogenes. Genomics 80(1):71–77
Van Kleunen M, Röckle M, Stift M (2015) Admixture between native and invasive populations may increase invasiveness of Mimulus guttatus. Proc R Soc Lond B 282:20151487
Wang L, Liu S, Zhuang Z, Guo L, Meng Z, Lin H (2013) Population genetic studies revealed local adaptation in a high gene-flow marine fish, the small yellow croaker (Larimichthys polyactis). PLoS ONE 8:e83493
Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN (2005) Barcoding Australia’s fish species. Philos Trans R Soc B 360:1847–1857
White DJ, Wolff JN, Pierson M, Gemmell NJ (2008) Revealing the hidden complexities of mtDNA inheritance. Mol Ecol 17:4925–4942
Whitfield PE, Hare JA, David AW, Harter SL, Munoz RC, Addison CM (2007) Abundance estimates of the Indo-Pacific lionfish Pterois volitans/miles complex in the Western North Atlantic. Biol Invasions 9:53–64
Wilcox CL, Motomura H, Matsunuma M, Bowen BW (2018) Phylogeography of lionfishes (Pterois) indicate taxonomic over splitting and hybrid origin of the invasive Pterois volitans. J Hered 109:162–175
Worm B, Barbier EB, Beaumont N, Duffy JE, Folke C, Halpern BS, Jackson JBC et al (2006) Impacts of biodiversity loss on ocean ecosystem services. Science 314:787–790
Yaakub SM, Bellwood DR, van Herwerden L, Walsh FM (2006) Hybridization in coral reef fishes: introgression and bi-directional gene exchange in Thalassoma (family Labridae). Mol Phylogenet Evol 40:84–100
Zapfe L, Freeland JR (2015) Heterosis in invasive F 1 cattail hybrids (Typha×glauca). Aquat Bot 125:44–47
Zieliński P, Nadachowska-Brzyska K, Wielstra B, Szkotak R, Covaciu-Marcov SD, Cogălniceanu D, Babik W (2013) No evidence for nuclear introgression despite complete mt DNA replacement in the Carpathian newt (L issotriton montandoni). Mol Ecol 22(7):1884–1903
Acknowledgements
We acknowledge and thank Amy Brower, Margaret Hunter, Kirsty McFarland, Captain Andrew Ross, Tyler Steube, Divers Supply (Jacksonville), Suncoast Spearfishing Challenge, Pensacola Lionfish Awareness & Removal Day, Florida Fish and Wildlife Conservation Commission, Upper Keys Lionfish Derby (Key Largo), and Forfar Field Station (Andros Island) for help in collection of tissue samples.
Author information
Authors and Affiliations
Contributions
AMJ conceived the research and coordinated sampling. Laboratory work, including DNA extractions and PCR, and subsequent analyses were performed by JW and reviewed by AMJ JW wrote the manuscript with significant review performed by AMJ.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10592_2020_1263_MOESM1_ESM.xlsx
Supplementary file1 (XLSX 21 kb). Appendix 1. Sample identification number, location and match when Saner sequence results were queried against BLASTn database. D-loop and COI sequences are identified in Genbank as P. volitans or P. miles, and S7 intron-1 sequences are identified as Indian Ocean lineage (representing P. miles), and Pacific Ocean lineage (representing P. volitans/russelli/lunulata)
10592_2020_1263_MOESM2_ESM.xlsx
Supplementary file2 (XLSX 98 kb). Appendix 2. Single nucleotide polymorphisms identified when aligned to the P. volitans reference, accession #KM488633.1
10592_2020_1263_MOESM3_ESM.xlsx
Supplementary file3 (XLSX 143 kb). Appendix 3. Single nucleotide polymorphisms identified when aligned to the P. miles reference, accession #K022697.1
Rights and permissions
About this article
Cite this article
Whitaker, J.M., Janosik, A.M. Missing the mark(er): pseudogenes identified through whole mitochondrial genome sequencing provide new insight into invasive lionfish genetics. Conserv Genet 21, 467–480 (2020). https://doi.org/10.1007/s10592-020-01263-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-020-01263-9